正在加载图片...
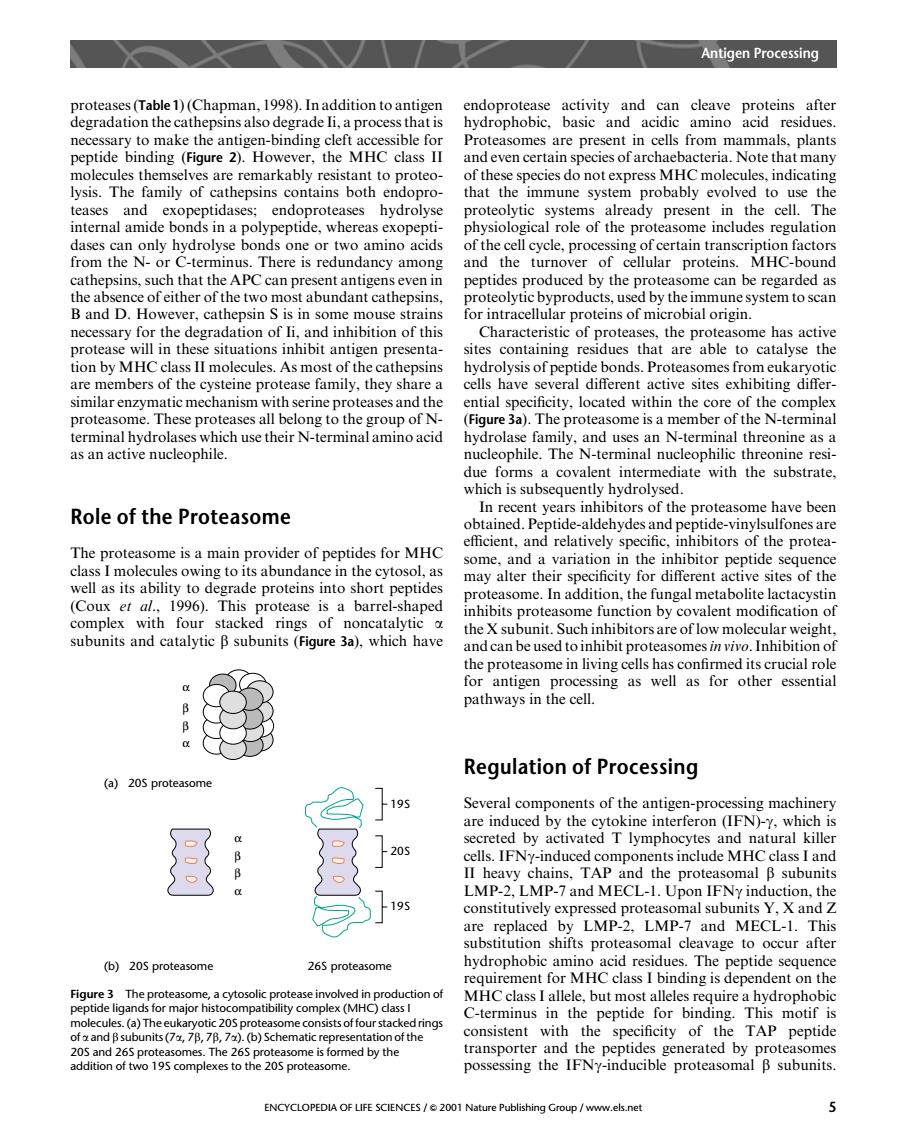
Antigen Processing prote rate )(Cha) endoprotease make the antis peptide binding (igure Hoeve the MHC Pro als nla and even certain species of archaebacteria.Note that man mily of hepsins bot immune syster proba to us dases can only hydrolyse bonds one or two amino acids of the cell cycle,processing of certain transcription factors s.There is redundancy among and the turnove of cellular proteins. MHC-bound peptides produced by the prote ome can be regarded as B and D.However.cathepsin s is in some mouse strains necessary for the degradation of li,and inhibition of this Characteristic of proteases,the proteasome has active situations inhibit antigen presenta sites containing that are able to catalyse the II molecule 5.As most of ond hibi cuka roteases and the ntial cificity.located within the core of the proteasome.These proteases all belong to the group of N (Figure 3a).The proteasome is a member of the N-terminal their N-erminal terminal hydrol nal threonine as a Role of the Proteasome In recent years inhibitors of the proteasome have been ptide aldehydesand pe de-vin he protea The proteasome is a main provider of peptides for MHC tots abundanc the cytos may alter their specificity for different active sites of the (Coux et al..1996).This ease is a barrel shaned easome.In addition,the fungal metabolite lactacystin alent subunits and catalytic B subunits(Fiqure 3a).which have the c the proteasome in living cells has confirmed its crucial rol for antigen processing as well as for other essential pathways in the cell. Regulation of Processing (a)205 proteasom 19 Several com n-proc secreted by activated T lymphoc ytes and natural killer 20 cells.IFNy-induced components include MHCclass I and ns,IA the protea oma B subunits -and M are replaced by LMP-2.LMP-7 and MECL-1.This substitution shifts proteasomal cleavage to occur after (b)205proteasome 265 proteasome hydrophobic amin Hd resi due peptid sequenc ing is Figure3 The proteasome,a cytosolic pro e involved in production of a)Th or majo C-terminus in the peptide for binding.This motif is consistent with the specificity of the TAP peptide and 265 pr The 265 on or tv 195 complexes to th ENCYCLOPEDIA OF LIFE SCIENCES/2001 Nature Publishing Group/www.els.net proteases (Table1) (Chapman, 1998). In addition to antigen degradation the cathepsins also degrade Ii, a process that is necessary to make the antigen-binding cleft accessible for peptide binding (Figure 2). However, the MHC class II molecules themselves are remarkably resistant to proteolysis. The family of cathepsins contains both endoproteases and exopeptidases; endoproteases hydrolyse internal amide bonds in a polypeptide, whereas exopeptidases can only hydrolyse bonds one or two amino acids from the N- or C-terminus. There is redundancy among cathepsins, such that the APC can present antigens even in the absence of either of the two most abundant cathepsins, B and D. However, cathepsin S is in some mouse strains necessary for the degradation of Ii, and inhibition of this protease will in these situations inhibit antigen presentation by MHC class II molecules. As most of the cathepsins are members of the cysteine protease family, they share a similar enzymatic mechanism with serine proteases and the proteasome. These proteases all belong to the group of Nterminal hydrolases which use their N-terminal amino acid as an active nucleophile. Role of the Proteasome The proteasome is a main provider of peptides for MHC class I molecules owing to its abundance in the cytosol, as well as its ability to degrade proteins into short peptides (Coux et al., 1996). This protease is a barrel-shaped complex with four stacked rings of noncatalytic a subunits and catalytic b subunits (Figure 3a), which have endoprotease activity and can cleave proteins after hydrophobic, basic and acidic amino acid residues. Proteasomes are present in cells from mammals, plants and even certain species of archaebacteria. Note that many of these species do not express MHC molecules, indicating that the immune system probably evolved to use the proteolytic systems already present in the cell. The physiological role of the proteasome includes regulation of the cell cycle, processing of certain transcription factors and the turnover of cellular proteins. MHC-bound peptides produced by the proteasome can be regarded as proteolytic byproducts, used by the immune system to scan for intracellular proteins of microbial origin. Characteristic of proteases, the proteasome has active sites containing residues that are able to catalyse the hydrolysis of peptide bonds. Proteasomes from eukaryotic cells have several different active sites exhibiting differential specificity, located within the core of the complex (Figure 3a). The proteasome is a member of the N-terminal hydrolase family, and uses an N-terminal threonine as a nucleophile. The N-terminal nucleophilic threonine residue forms a covalent intermediate with the substrate, which is subsequently hydrolysed. In recent years inhibitors of the proteasome have been obtained. Peptide-aldehydes and peptide-vinylsulfones are efficient, and relatively specific, inhibitors of the proteasome, and a variation in the inhibitor peptide sequence may alter their specificity for different active sites of the proteasome. In addition, the fungal metabolite lactacystin inhibits proteasome function by covalent modification of the X subunit. Such inhibitors are of low molecular weight, and can be used to inhibit proteasomesin vivo. Inhibition of the proteasome in living cells has confirmed its crucial role for antigen processing as well as for other essential pathways in the cell. Regulation of Processing Several components of the antigen-processing machinery are induced by the cytokine interferon (IFN)-g, which is secreted by activated T lymphocytes and natural killer cells. IFNg-induced components include MHC class I and II heavy chains, TAP and the proteasomal b subunits LMP-2, LMP-7 and MECL-1. Upon IFNg induction, the constitutively expressed proteasomal subunits Y, X and Z are replaced by LMP-2, LMP-7 and MECL-1. This substitution shifts proteasomal cleavage to occur after hydrophobic amino acid residues. The peptide sequence requirement for MHC class I binding is dependent on the MHC class I allele, but most alleles require a hydrophobic C-terminus in the peptide for binding. This motif is consistent with the specificity of the TAP peptide transporter and the peptides generated by proteasomes possessing the IFNg-inducible proteasomal b subunits. α β β α (a) 20S proteasome α β β α (b) 20S proteasome 26S proteasome 19S 20S 19S Figure 3 The proteasome, a cytosolic protease involved in production of peptide ligands for major histocompatibility complex (MHC) class I molecules. (a) The eukaryotic 20S proteasome consists of four stacked rings of a and b subunits (7a, 7b, 7b, 7a). (b) Schematic representation of the 20S and 26S proteasomes. The 26S proteasome is formed by the addition of two 19S complexes to the 20S proteasome. Antigen Processing ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group / www.els.net 5