正在加载图片...
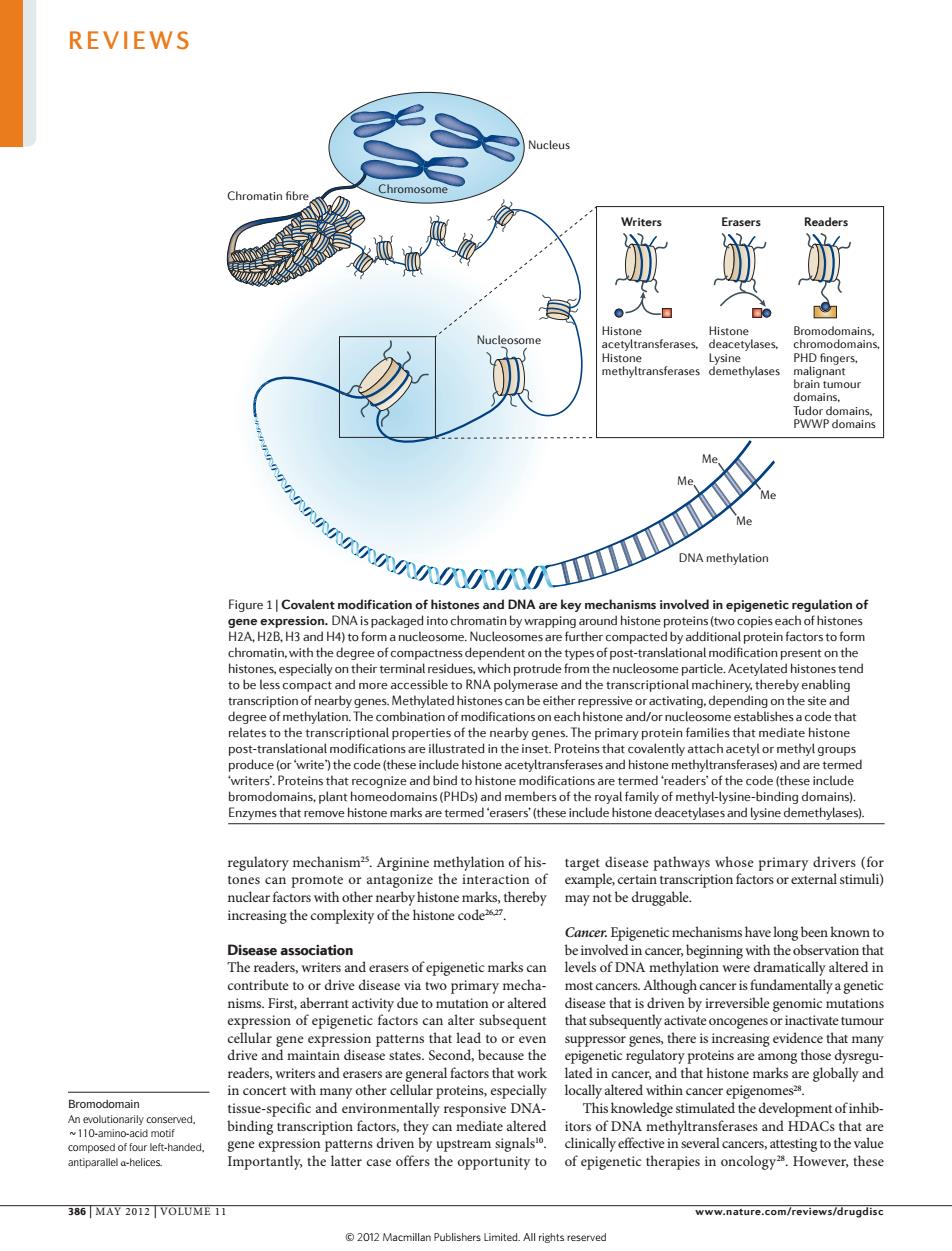
REVIEWS dorpaomin //y igure 1 Covale n of his d in epige eachaationo .H2B.H oteinfactostoioe tivating.dep on the site al prope ry prote diate histone the cod e acety ases and histone m and an pla s)and m ers of th of methyl-lysir ng domains the inte been knownt evels of DNA e dramatically altered ir most cr ougn cancer is amenta ssion of epigenetic factors n alter subsequent that inactivatetumou in canc This kno ent ofinhib. the latter case offers the opportunity to 386 MAY 2012 VOLUME 11 www.nature.com/reviews/drugdisc 2012 Macmillan Publishers Limited.All riehts reserved Nucleus Writers Erasers Readers Nature Reviews | Drug Discovery Histone acetyltransferases, Histone methyltransferases Histone deacetylases, Lysine demethylases Bromodomains, chromodomains, PHD fingers, malignant brain tumour domains, Tudor domains, PWWP domains DNA methylation Me Me Me Me Chromatin fibre Chromosome Nucleosome Bromodomain An evolutionarily conserved, ~110-amino-acid motif composed of four left-handed, antiparallel α-helices. regulatory mechanism25. Arginine methylation of histones can promote or antagonize the interaction of nuclear factors with other nearby histone marks, thereby increasing the complexity of the histone code26,27. Disease association The readers, writers and erasers of epigenetic marks can contribute to or drive disease via two primary mechanisms. First, aberrant activity due to mutation or altered expression of epigenetic factors can alter subsequent cellular gene expression patterns that lead to or even drive and maintain disease states. Second, because the readers, writers and erasers are general factors that work in concert with many other cellular proteins, especially tissue-specific and environmentally responsive DNAbinding transcription factors, they can mediate altered gene expression patterns driven by upstream signals10. Importantly, the latter case offers the opportunity to target disease pathways whose primary drivers (for example, certain transcription factors or external stimuli) may not be druggable. Cancer. Epigenetic mechanisms have long been known to be involved in cancer, beginning with the observation that levels of DNA methylation were dramatically altered in most cancers. Although cancer is fundamentally a genetic disease that is driven by irreversible genomic mutations that subsequently activate oncogenes or inactivate tumour suppressor genes, there is increasing evidence that many epigenetic regulatory proteins are among those dysregulated in cancer, and that histone marks are globally and locally altered within cancer epigenomes28. This knowledge stimulated the development of inhibitors of DNA methyltransferases and HDACs that are clinically effective in several cancers, attesting to the value of epigenetic therapies in oncology28. However, these Figure 1 | Covalent modification of histones and DNA are key mechanisms involved in epigenetic regulation of gene expression. DNA is packaged into chromatin by wrapping around histone proteins (two copies each of histones H2A, H2B, H3 and H4) to form a nucleosome. Nucleosomes are further compacted by additional protein factors to form chromatin, with the degree of compactness dependent on the types of post-translational modification present on the histones, especially on their terminal residues, which protrude from the nucleosome particle. Acetylated histones tend to be less compact and more accessible to RNA polymerase and the transcriptional machinery, thereby enabling transcription of nearby genes. Methylated histones can be either repressive or activating, depending on the site and degree of methylation. The combination of modifications on each histone and/or nucleosome establishes a code that relates to the transcriptional properties of the nearby genes. The primary protein families that mediate histone post-translational modifications are illustrated in the inset. Proteins that covalently attach acetyl or methyl groups produce (or ‘write’) the code (these include histone acetyltransferases and histone methyltransferases) and are termed ‘writers’. Proteins that recognize and bind to histone modifications are termed ‘readers’ of the code (these include bromodomains, plant homeodomains (PHDs) and members of the royal family of methyl-lysine-binding domains). Enzymes that remove histone marks are termed ‘erasers’ (these include histone deacetylases and lysine demethylases). REVIEWS 386 | MAY 2012 | VOLUME 11 www.nature.com/reviews/drugdisc © 2012 Macmillan Publishers Limited. All rights reserved