正在加载图片...
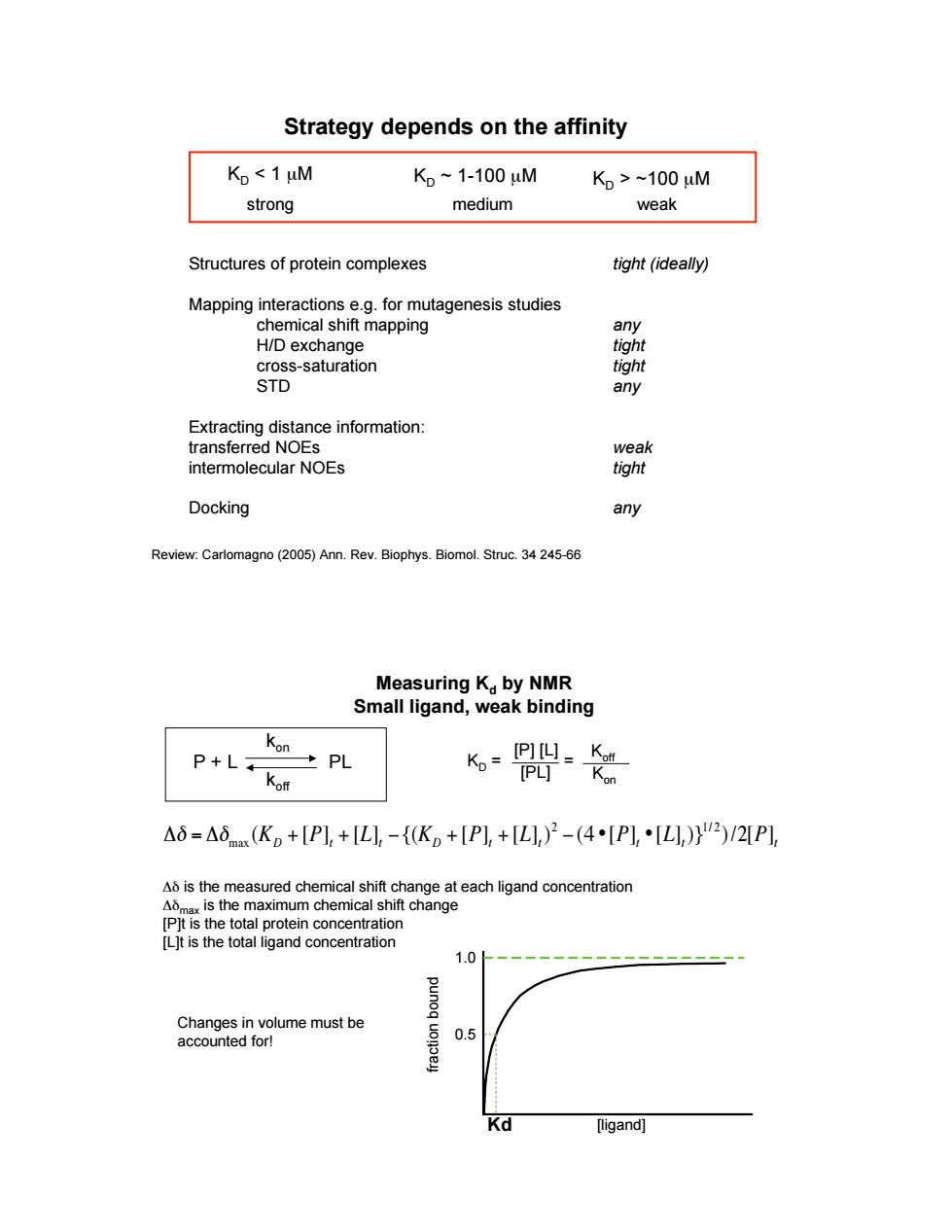
Strategy depends on the affinity Ko 1uM K,~1-100uM K,>~100uM strong medium weak Structures of protein complexes tight(ideally) Mapping interactions e.g.for mutagenesis studies chemical shift mapping H/D exchange cross-saturation tight STD any Extracting distance information: transferred NOEs intermolecular NOEs lon Docking any Review:Carlomagno(2005)Ann.Rev.Biophys.Biomol.Struc.34245-66 Measuring Ka by NMR Small ligand,weak binding P+L K聘 △6=△6(K。+[P1,+L,-{K。+[P1,+L,)2-(4[P,·[,)}Ψ2)/2P1 A is the measured chemical shift change at each ligand concentration t change Lt is the total igand conc [ligand]tight (ideally) any tight tight any weak tight any Structures of protein complexes Mapping interactions e.g. for mutagenesis studies chemical shift mapping H/D exchange cross-saturation STD Extracting distance information: transferred NOEs intermolecular NOEs Docking Strategy depends on the affinity KD < 1 µM KD ~ 1-100 µM KD > ~100 µM strong medium weak Review: Carlomagno (2005) Ann. Rev. Biophys. Biomol. Struc. 34 245-66 Measuring Kd by NMR Small ligand, weak binding ! "# = "#max (KD + [P]t + [L]t ${(KD + [P]t + [L]t) 2 $ (4 •[P]t •[L]t)}1/ 2 )/2[P]t #$ is the measured chemical shift change at each ligand concentration #$max is the maximum chemical shift change [P]t is the total protein concentration [L]t is the total ligand concentration [ligand] 1.0 0.5 f r a ctio n b o u n d Kd P + L PL kon koff KD = [P] [L] [PL] = Koff Kon Changes in volume must be accounted for!