正在加载图片...
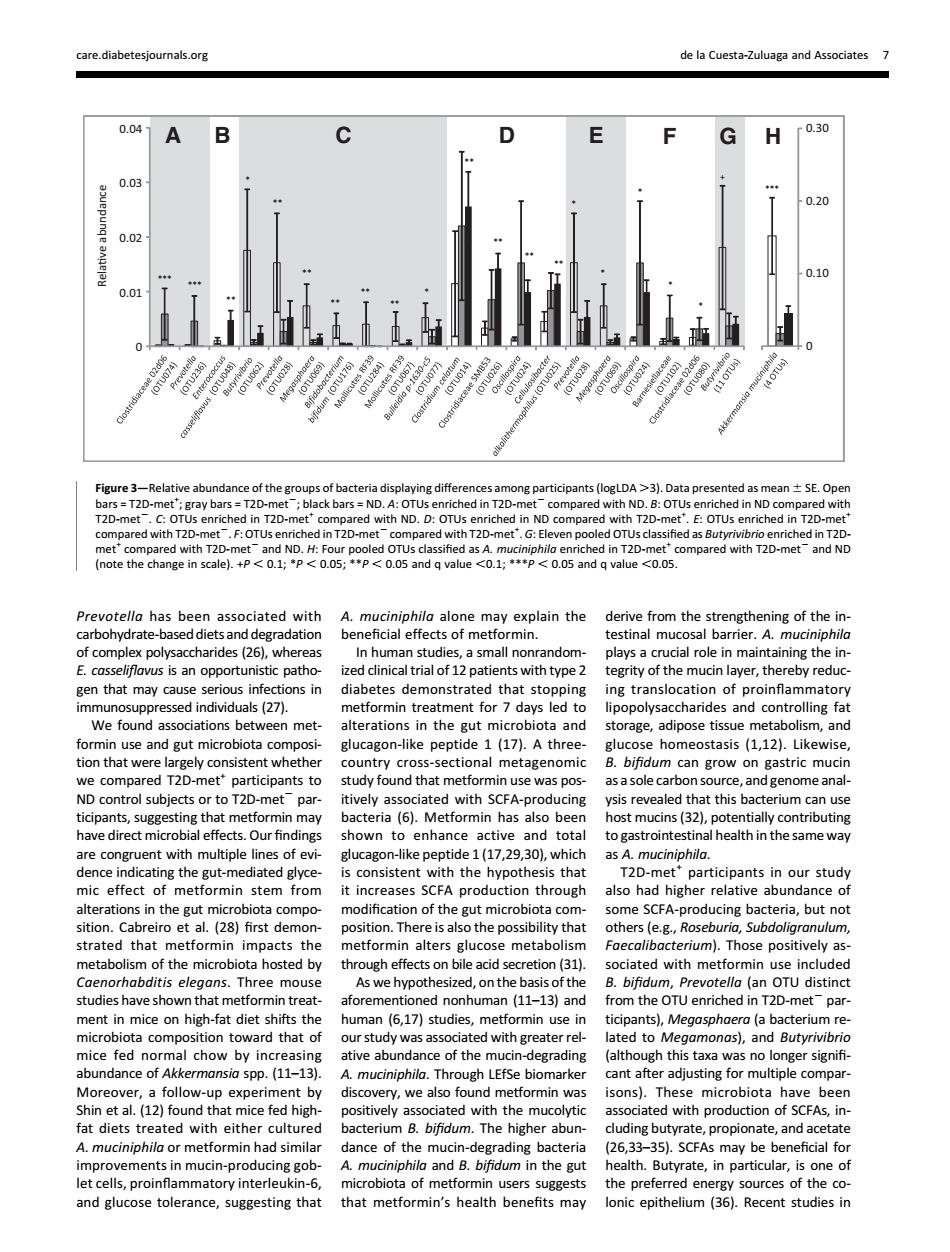
care diabetesio als.org de la Cuesta-Zuluaga and Associates 7 0.04 A G H f0.30 0.2 0.1 0.0 -Re of th ng diff ts (logLDA >3).Data p nted n±sE.Ope with ND.D 20- 2-met and NE Prevotello has been associated with A.muciniphila alone may explain the derive from the strengthening of the in carbohydrate degradation beneficial effects of m rmin. estinal mucosal barrie terit of the mucin laver.ther gen that may cause se tions in diabetes demonstrated that stopping ing translocation of proinfammatory 527 ormin treat da ipopolys formin use and eut microbiota com glucag on-like peptide 1(17).A three gucose homeostasis (112).Likewise clmtoCioieiectoalimctagenoic r to T2D s p with SCEA t this bad may bacteria(6).Metformin has also been potentially contributing nce al health in the same way T2D-metparticipants in our study mic effect of metfo min stem from it increases SCFA production through also had higher relative abundance o sition on of t e gut microb ing bac na,but not impacts the metformin alters glucose metabolism metabolism f the microbl ota hosted by through effec on bile aci ( studies have shown that met aforeme uman (11-13)and ment in mice on high-fat diet shifts the human (6,17)studies,metformin use in ticipants),Megasphoera (a bac com rd that o b dy was n g ater re nd Butyri LEfSe biom overy,we also 50n5. mi robiota ave beer fat diets ed with aith rate A.muciniphila or metformin had simila (26,33-35).SCFAs may be beneficial for mprovements in mu dance of the mucin-eading bacteri In-pre and Butyrate,in particu one and glucose tolerance,suggesting that that metformin's health henefits mav lonic epithelium (35).Recent studies inPrevotella has been associated with carbohydrate-based diets and degradation of complex polysaccharides (26), whereas E. casseliflavus is an opportunistic pathogen that may cause serious infections in immunosuppressed individuals (27). We found associations between metformin use and gut microbiota composition that were largely consistent whether we compared T2D-met+ participants to ND control subjects or to T2D-met2 participants, suggesting that metformin may have direct microbial effects. Our findings are congruent with multiple lines of evidence indicating the gut-mediated glycemic effect of metformin stem from alterations in the gut microbiota composition. Cabreiro et al. (28) first demonstrated that metformin impacts the metabolism of the microbiota hosted by Caenorhabditis elegans. Three mouse studies have shown that metformin treatment in mice on high-fat diet shifts the microbiota composition toward that of mice fed normal chow by increasing abundance of Akkermansia spp. (11–13). Moreover, a follow-up experiment by Shin et al. (12) found that mice fed highfat diets treated with either cultured A. muciniphila or metformin had similar improvements in mucin-producing goblet cells, proinflammatory interleukin-6, and glucose tolerance, suggesting that A. muciniphila alone may explain the beneficial effects of metformin. In human studies, a small nonrandomized clinical trial of 12 patients with type 2 diabetes demonstrated that stopping metformin treatment for 7 days led to alterations in the gut microbiota and glucagon-like peptide 1 (17). A threecountry cross-sectional metagenomic study found that metformin use was positively associated with SCFA-producing bacteria (6). Metformin has also been shown to enhance active and total glucagon-like peptide 1 (17,29,30), which is consistent with the hypothesis that it increases SCFA production through modification of the gut microbiota composition. There is also the possibility that metformin alters glucose metabolism through effects on bile acid secretion (31). As we hypothesized, on the basis of the aforementioned nonhuman (11–13) and human (6,17) studies, metformin use in our study was associated with greater relative abundance of the mucin-degrading A. muciniphila. Through LEfSe biomarker discovery, we also found metformin was positively associated with the mucolytic bacterium B. bifidum. The higher abundance of the mucin-degrading bacteria A. muciniphila and B. bifidum in the gut microbiota of metformin users suggests that metformin’s health benefits may derive from the strengthening of the intestinal mucosal barrier. A. muciniphila plays a crucial role in maintaining the integrity of the mucin layer, thereby reducing translocation of proinflammatory lipopolysaccharides and controlling fat storage, adipose tissue metabolism, and glucose homeostasis (1,12). Likewise, B. bifidum can grow on gastric mucin as a sole carbon source, and genome analysis revealed that this bacterium can use host mucins (32), potentially contributing to gastrointestinal health in the same way as A. muciniphila. T2D-met+ participants in our study also had higher relative abundance of some SCFA-producing bacteria, but not others (e.g., Roseburia, Subdoligranulum, Faecalibacterium). Those positively associated with metformin use included B. bifidum, Prevotella (an OTU distinct from the OTU enriched in T2D-met2 participants), Megasphaera (a bacterium related to Megamonas), and Butyrivibrio (although this taxa was no longer signifi- cant after adjusting for multiple comparisons). These microbiota have been associated with production of SCFAs, including butyrate, propionate, and acetate (26,33–35). SCFAs may be beneficial for health. Butyrate, in particular, is one of the preferred energy sources of the colonic epithelium (36). Recent studies in 0 0.01 0.02 0.03 0.04 Relave abundance 0 0.10 0.20 0.30 AB C D E F GH *** *** ** * ** ** ** ** ** * ** ** ** ** + *** * * * * * Figure 3—Relative abundance of the groups of bacteria displaying differences among participants (logLDA .3). Data presented as mean 6 SE. Open bars = T2D-met+ ; gray bars = T2D-met2; black bars = ND. A: OTUs enriched in T2D-met2 compared with ND. B: OTUs enriched in ND compared with T2D-met2. C: OTUs enriched in T2D-met+ compared with ND. D: OTUs enriched in ND compared with T2D-met+ . E: OTUs enriched in T2D-met+ compared with T2D-met2. F: OTUs enriched in T2D-met2 compared with T2D-met+ . G: Eleven pooled OTUs classified as Butyrivibrio enriched in T2Dmet+ compared with T2D-met2 and ND. H: Four pooled OTUs classified as A. muciniphila enriched in T2D-met+ compared with T2D-met2 and ND (note the change in scale). +P , 0.1; *P , 0.05; **P , 0.05 and q value ,0.1; ***P , 0.05 and q value ,0.05. care.diabetesjournals.org de la Cuesta-Zuluaga and Associates 7�