正在加载图片...
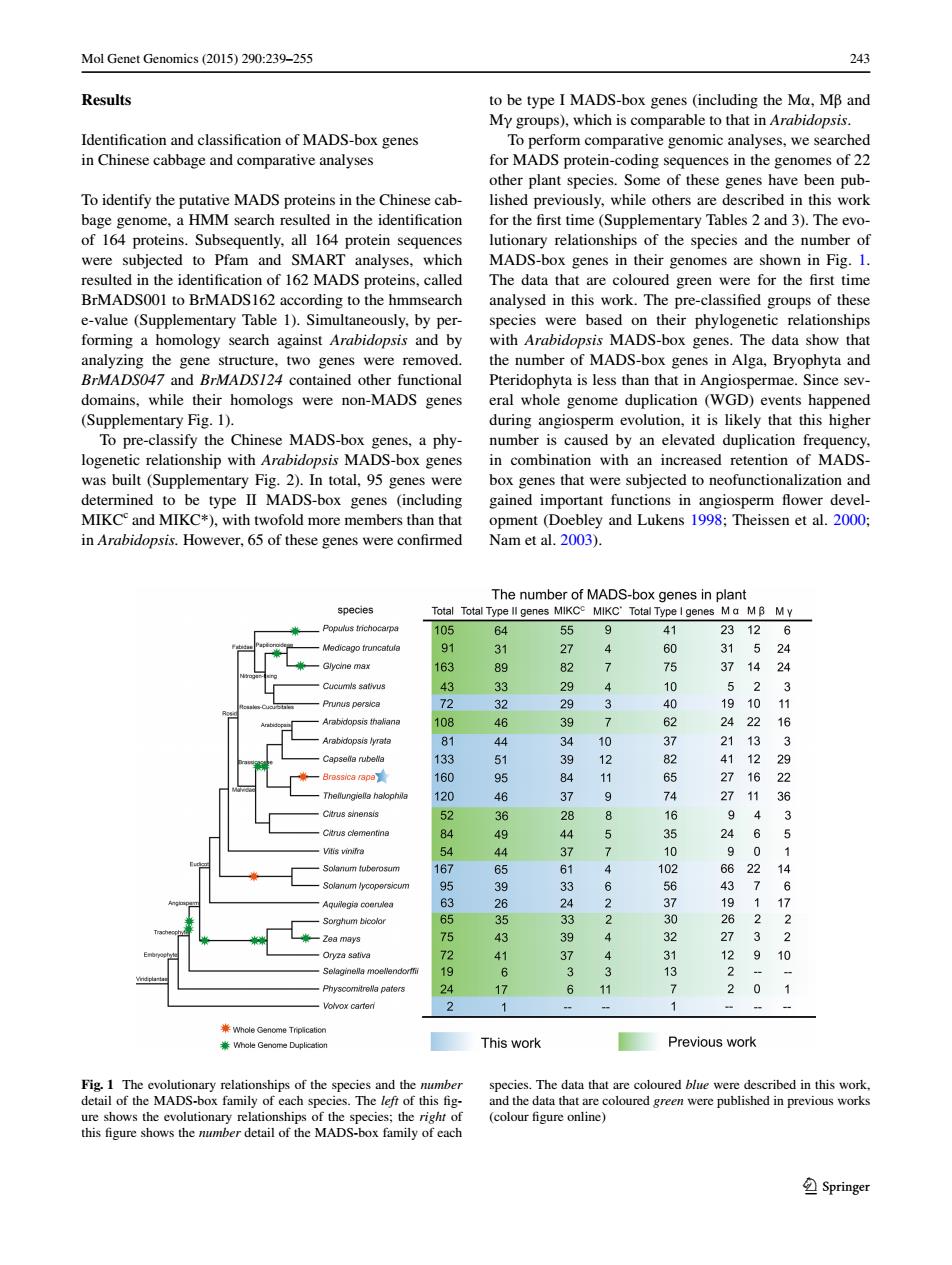
Mol Genet Genomics(2015)290:239-255 243 Results to be type I MADS-box genes (including the Ma,MB and My groups),which is comparable to that in Arabidopsis. Identification and classification of MADS-box genes To perform comparative genomic analyses,we searched in Chinese cabbage and comparative analyses for MADS protein-coding sequences in the genomes of 22 other plant species.Some of these genes have been pub- To identify the putative MADS proteins in the Chinese cab- lished previously,while others are described in this work bage genome,a HMM search resulted in the identification for the first time(Supplementary Tables 2 and 3).The evo- of 164 proteins.Subsequently,all 164 protein sequences lutionary relationships of the species and the number of were subjected to Pfam and SMART analyses,which MADS-box genes in their genomes are shown in Fig.1. resulted in the identification of 162 MADS proteins,called The data that are coloured green were for the first time BrMADS001 to BrMADS162 according to the hmmsearch analysed in this work.The pre-classified groups of these e-value (Supplementary Table 1).Simultaneously,by per- species were based on their phylogenetic relationships forming a homology search against Arabidopsis and by with Arabidopsis MADS-box genes.The data show that analyzing the gene structure,two genes were removed. the number of MADS-box genes in Alga,Bryophyta and BrMADS047 and BrMADS124 contained other functional Pteridophyta is less than that in Angiospermae.Since sev- domains,while their homologs were non-MADS genes eral whole genome duplication(WGD)events happened (Supplementary Fig.1). during angiosperm evolution,it is likely that this higher To pre-classify the Chinese MADS-box genes,a phy- number is caused by an elevated duplication frequency, logenetic relationship with Arabidopsis MADS-box genes in combination with an increased retention of MADS- was built (Supplementary Fig.2).In total,95 genes were box genes that were subjected to neofunctionalization and determined to be type II MADS-box genes (including gained important functions in angiosperm flower devel- MIKC and MIKC*),with twofold more members than that opment(Doebley and Lukens 1998;Theissen et al.2000; in Arabidopsis.However,65 of these genes were confirmed Nam et al.2003). The number of MADS-box genes in plant species Total Total Type ll genes MIKCC MIKC'Total Type I genes Ma MB My Populus trichocarpa 105 64 55 9 41 23126 Medicago fruncatufa 91 31 27 60 31524 -Glycine max 163 89 82 7 75 3714 品 Cucimls sativus 3 4 52 72 32 29 3 40 1910 Arabidopsis thaliana 108 46 3 > 24 22 16 Arabidopsis lyrata 81 44 10 21 13 3 Caosele rubela 133 9 8 4112 160 95 1 16 2 Theiungielle halophia 120 46 9 7 27 11 36 52 36 2 8 9 3 Citnis clementine 84 9 24 54 4 3 10 167 4 1 66 22 14 Solanum lycopersicu 95 6 56 7 6 63 2 1 Sorghum bicola 65 33 30 Zea mays 75 43 4 27 2 Oryza sativa 72 37 4 31 12 9 Selaginella moellendorfi 19 6 3 13 3 24 6 11 2 0 Volvox carten 2 Whole Genome Tripiication Whole Genome Duplication This work Previous work Fig.1 The evolutionary relationships of the species and the number species.The data that are coloured blue were described in this work. detail of the MADS-box family of each species.The left of this fig- and the data that are coloured green were published in previous works ure shows the evolutionary relationships of the species;the right of (colour figure online) this figure shows the number detail of the MADS-box family of each ②SpringerMol Genet Genomics (2015) 290:239–255 243 1 3 Results Identification and classification of MADS-box genes in Chinese cabbage and comparative analyses To identify the putative MADS proteins in the Chinese cabbage genome, a HMM search resulted in the identification of 164 proteins. Subsequently, all 164 protein sequences were subjected to Pfam and SMART analyses, which resulted in the identification of 162 MADS proteins, called BrMADS001 to BrMADS162 according to the hmmsearch e-value (Supplementary Table 1). Simultaneously, by performing a homology search against Arabidopsis and by analyzing the gene structure, two genes were removed. BrMADS047 and BrMADS124 contained other functional domains, while their homologs were non-MADS genes (Supplementary Fig. 1). To pre-classify the Chinese MADS-box genes, a phylogenetic relationship with Arabidopsis MADS-box genes was built (Supplementary Fig. 2). In total, 95 genes were determined to be type II MADS-box genes (including MIKCc and MIKC*), with twofold more members than that in Arabidopsis. However, 65 of these genes were confirmed to be type I MADS-box genes (including the Mα, Mβ and Mγ groups), which is comparable to that in Arabidopsis. To perform comparative genomic analyses, we searched for MADS protein-coding sequences in the genomes of 22 other plant species. Some of these genes have been published previously, while others are described in this work for the first time (Supplementary Tables 2 and 3). The evolutionary relationships of the species and the number of MADS-box genes in their genomes are shown in Fig. 1. The data that are coloured green were for the first time analysed in this work. The pre-classified groups of these species were based on their phylogenetic relationships with Arabidopsis MADS-box genes. The data show that the number of MADS-box genes in Alga, Bryophyta and Pteridophyta is less than that in Angiospermae. Since several whole genome duplication (WGD) events happened during angiosperm evolution, it is likely that this higher number is caused by an elevated duplication frequency, in combination with an increased retention of MADSbox genes that were subjected to neofunctionalization and gained important functions in angiosperm flower development (Doebley and Lukens 1998; Theissen et al. 2000; Nam et al. 2003). Fig. 1 The evolutionary relationships of the species and the number detail of the MADS-box family of each species. The left of this figure shows the evolutionary relationships of the species; the right of this figure shows the number detail of the MADS-box family of each species. The data that are coloured blue were described in this work, and the data that are coloured green were published in previous works (colour figure online)