正在加载图片...
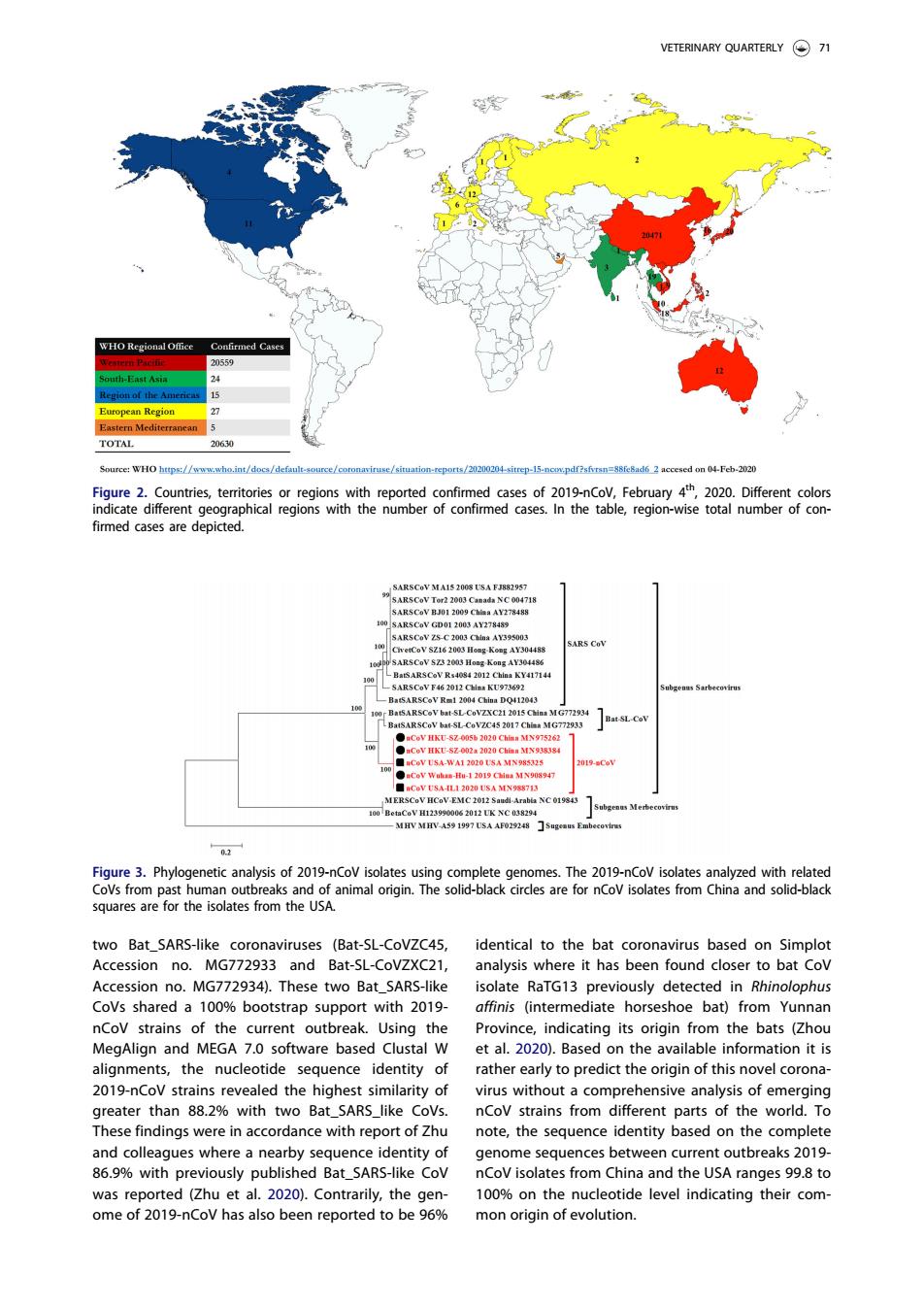
VETERINARY QUARTERLY(⊙刀 med Cases igure 2. ted confim edepicted. of 2019-nCo two Bat_SARS-like coronaviruses (Bat-SL-CoVZC45, identical to the bat coronavirus based on Simplot Accession no. MG772933 and Bat-SL-CoVZXC2 analysis where it has been found closer to bat Cov Accession no.MG772934).These two Bat_SARS-like solate RaTG13 previously detected in Rhinolophus otstrap s oport with 2015 norse noe ba ron na Pd MEGA 7O the ba alignments,the nucleotide sequence identity of ather early to r predict the orioin of this novelcoron 2019-nCov strains revealed the highest similarity of virus without a comprehensive analysis of emerging greater than 88.2%with two Bat_SARS_like Covs. nCov strains from different parts of the world.To hese findings were in accordance with report of Zhu note,the sequence identity based on the complete genome sequences between current outbreaks 2019 Bat_SARS V I eniepoeaobe96s evel two Bat_SARS-like coronaviruses (Bat-SL-CoVZC45, Accession no. MG772933 and Bat-SL-CoVZXC21, Accession no. MG772934). These two Bat_SARS-like CoVs shared a 100% bootstrap support with 2019- nCoV strains of the current outbreak. Using the MegAlign and MEGA 7.0 software based Clustal W alignments, the nucleotide sequence identity of 2019-nCoV strains revealed the highest similarity of greater than 88.2% with two Bat_SARS_like CoVs. These findings were in accordance with report of Zhu and colleagues where a nearby sequence identity of 86.9% with previously published Bat_SARS-like CoV was reported (Zhu et al. 2020). Contrarily, the genome of 2019-nCoV has also been reported to be 96% identical to the bat coronavirus based on Simplot analysis where it has been found closer to bat CoV isolate RaTG13 previously detected in Rhinolophus affinis (intermediate horseshoe bat) from Yunnan Province, indicating its origin from the bats (Zhou et al. 2020). Based on the available information it is rather early to predict the origin of this novel coronavirus without a comprehensive analysis of emerging nCoV strains from different parts of the world. To note, the sequence identity based on the complete genome sequences between current outbreaks 2019- nCoV isolates from China and the USA ranges 99.8 to 100% on the nucleotide level indicating their common origin of evolution. Figure 2. Countries, territories or regions with reported confirmed cases of 2019-nCoV, February 4th, 2020. Different colors indicate different geographical regions with the number of confirmed cases. In the table, region-wise total number of confirmed cases are depicted. Figure 3. Phylogenetic analysis of 2019-nCoV isolates using complete genomes. The 2019-nCoV isolates analyzed with related CoVs from past human outbreaks and of animal origin. The solid-black circles are for nCoV isolates from China and solid-black squares are for the isolates from the USA. VETERINARY QUARTERLY 71