正在加载图片...
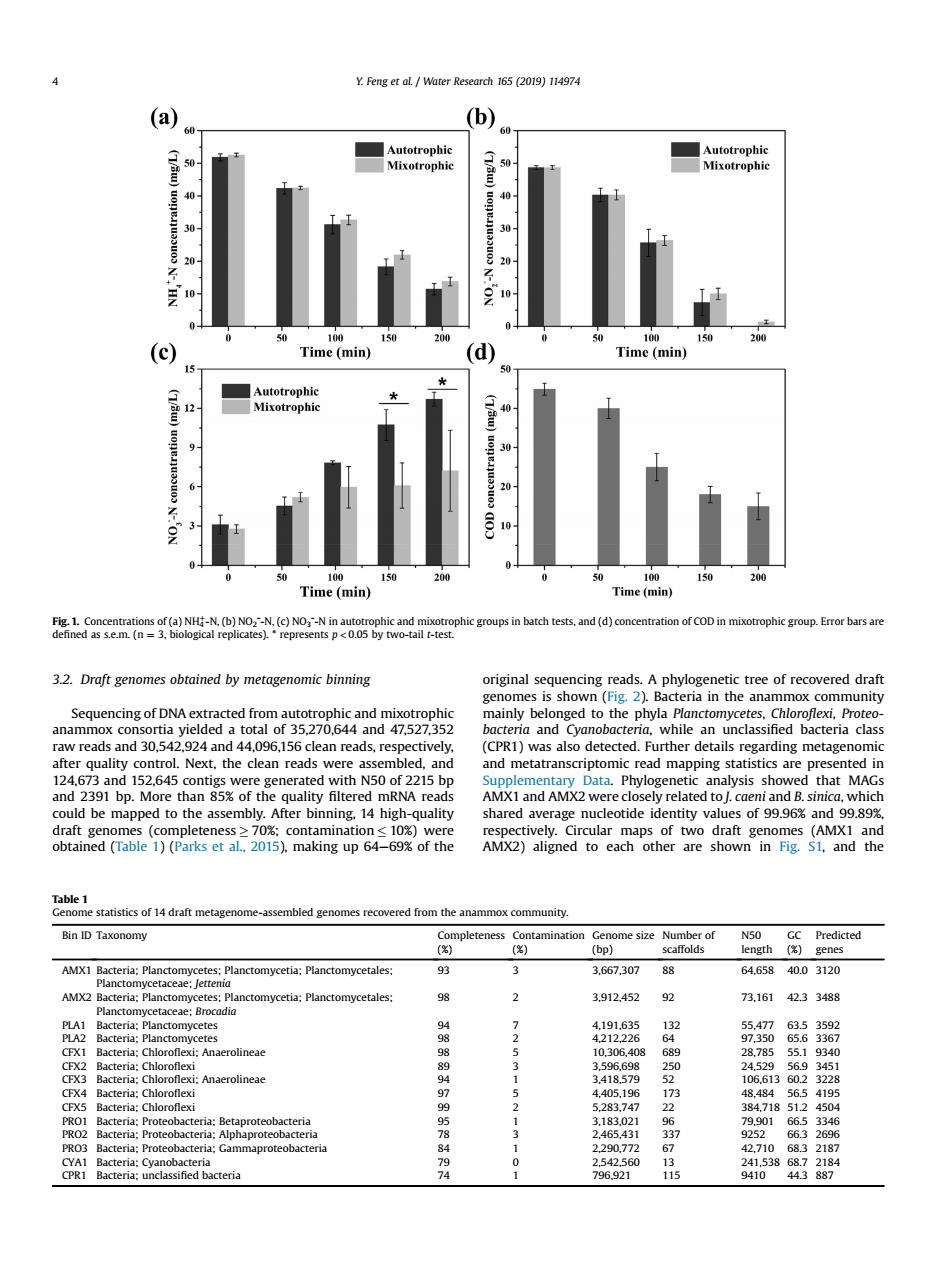
(b) 30 Time (min) (d) Time (min) Time(min) Time (mi cration of COD in ophic group.Error bars are 3.2.Draft genomes obtained by metagenomic binning original sequencing reads.A phylogenetic tree of recovered draft gen s sho d to the ria in the an nd3052gclded obacte while an bacteria clas d ir n≤10% f999 red from the anammox community. Bin ID Taxonomy 3.667.30788 64.6584003120 2 3912452 7.161 23 48 3 22907 796921 4103.2. Draft genomes obtained by metagenomic binning Sequencing of DNA extracted from autotrophic and mixotrophic anammox consortia yielded a total of 35,270,644 and 47,527,352 raw reads and 30,542,924 and 44,096,156 clean reads, respectively, after quality control. Next, the clean reads were assembled, and 124,673 and 152,645 contigs were generated with N50 of 2215 bp and 2391 bp. More than 85% of the quality filtered mRNA reads could be mapped to the assembly. After binning, 14 high-quality draft genomes (completeness 70%; contamination 10%) were obtained (Table 1) (Parks et al., 2015), making up 64e69% of the original sequencing reads. A phylogenetic tree of recovered draft genomes is shown (Fig. 2). Bacteria in the anammox community mainly belonged to the phyla Planctomycetes, Chloroflexi, Proteobacteria and Cyanobacteria, while an unclassified bacteria class (CPR1) was also detected. Further details regarding metagenomic and metatranscriptomic read mapping statistics are presented in Supplementary Data. Phylogenetic analysis showed that MAGs AMX1 and AMX2 were closely related to J. caeni and B. sinica, which shared average nucleotide identity values of 99.96% and 99.89%, respectively. Circular maps of two draft genomes (AMX1 and AMX2) aligned to each other are shown in Fig. S1, and the Fig. 1. Concentrations of (a) NH4 þ-N, (b) NO2ˉ-N, (c) NO3ˉ-N in autotrophic and mixotrophic groups in batch tests, and (d) concentration of COD in mixotrophic group. Error bars are defined as s.e.m. (n ¼ 3, biological replicates). * represents p < 0.05 by two-tail t-test. Table 1 Genome statistics of 14 draft metagenome-assembled genomes recovered from the anammox community. Bin ID Taxonomy Completeness (%) Contamination (%) Genome size (bp) Number of scaffolds N50 length GC (%) Predicted genes AMX1 Bacteria; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Jettenia 93 3 3,667,307 88 64,658 40.0 3120 AMX2 Bacteria; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Brocadia 98 2 3,912,452 92 73,161 42.3 3488 PLA1 Bacteria; Planctomycetes 94 7 4,191,635 132 55,477 63.5 3592 PLA2 Bacteria; Planctomycetes 98 2 4,212,226 64 97,350 65.6 3367 CFX1 Bacteria; Chloroflexi; Anaerolineae 98 5 10,306,408 689 28,785 55.1 9340 CFX2 Bacteria; Chloroflexi 89 3 3,596,698 250 24,529 56.9 3451 CFX3 Bacteria; Chloroflexi; Anaerolineae 94 1 3,418,579 52 106,613 60.2 3228 CFX4 Bacteria; Chloroflexi 97 5 4,405,196 173 48,484 56.5 4195 CFX5 Bacteria; Chloroflexi 99 2 5,283,747 22 384,718 51.2 4504 PRO1 Bacteria; Proteobacteria; Betaproteobacteria 95 1 3,183,021 96 79,901 66.5 3346 PRO2 Bacteria; Proteobacteria; Alphaproteobacteria 78 3 2,465,431 337 9252 66.3 2696 PRO3 Bacteria; Proteobacteria; Gammaproteobacteria 84 1 2,290,772 67 42,710 68.3 2187 CYA1 Bacteria; Cyanobacteria 79 0 2,542,560 13 241,538 68.7 2184 CPR1 Bacteria; unclassified bacteria 74 1 796,921 115 9410 44.3 887 4 Y. Feng et al. / Water Research 165 (2019) 114974