正在加载图片...
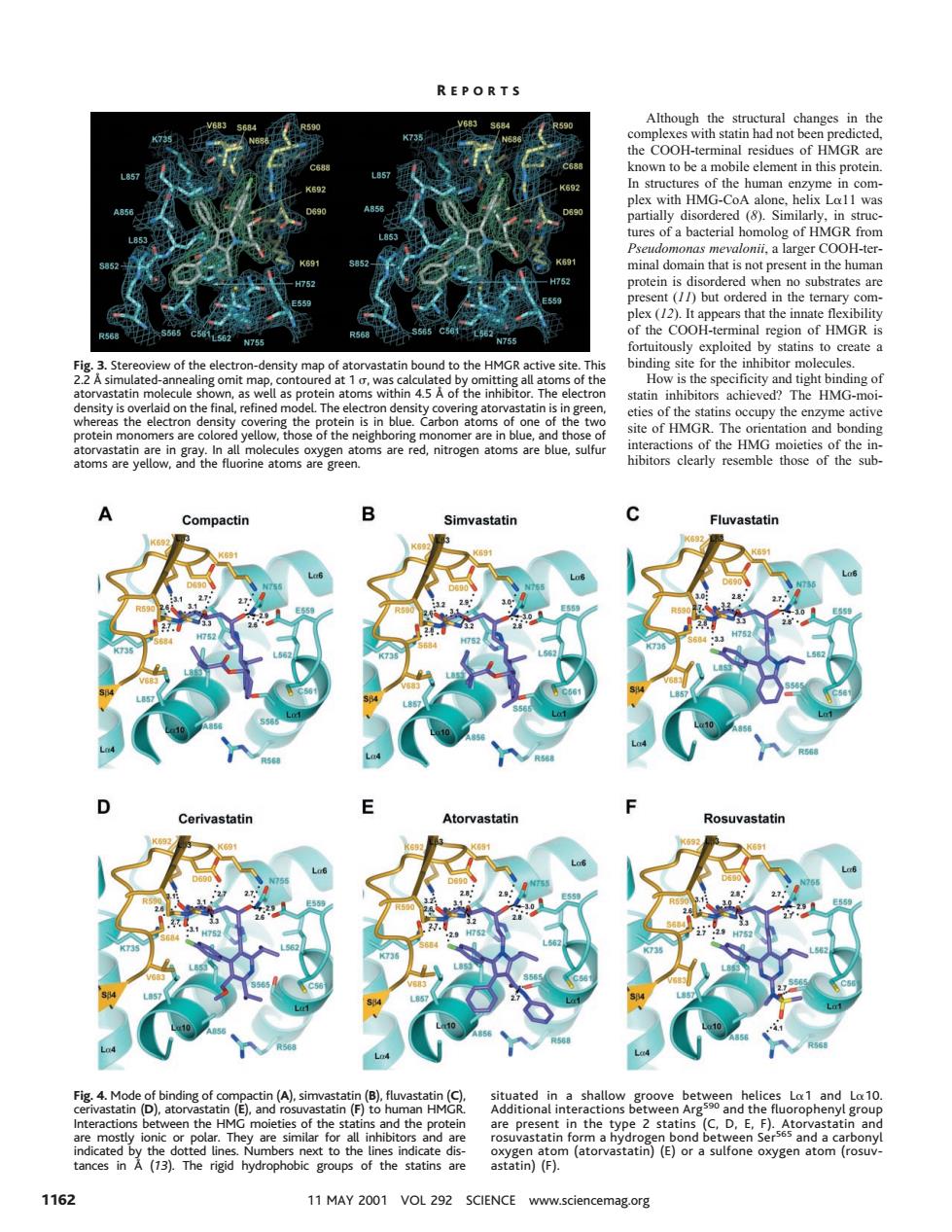
REPORTS Although the structural changes in the known to be a m element in this proteir with HMG was ures of a bacterial homolog of HMGR from in the human appears that loited by statins to create a sity map ng sit tight 0 chie HM f the h are red,nitrogen atoms are blue,sulfu toms are vellow and the fluorine atoms are gree ibitors clarly resemble those of the sub vastatin Cerivastatin d the n the Atorvastati astatin)(F). 1162 11MAY2001VOL292 SCIENCE www.sciencemag.org Although the structural changes in the complexes with statin had not been predicted, the COOH-terminal residues of HMGR are known to be a mobile element in this protein. In structures of the human enzyme in complex with HMG-CoA alone, helix La11 was partially disordered (8). Similarly, in structures of a bacterial homolog of HMGR from Pseudomonas mevalonii, a larger COOH-terminal domain that is not present in the human protein is disordered when no substrates are present (11) but ordered in the ternary complex (12). It appears that the innate flexibility of the COOH-terminal region of HMGR is fortuitously exploited by statins to create a binding site for the inhibitor molecules. How is the specificity and tight binding of statin inhibitors achieved? The HMG-moieties of the statins occupy the enzyme active site of HMGR. The orientation and bonding interactions of the HMG moieties of the inhibitors clearly resemble those of the subFig. 3. Stereoview of the electron-density map of atorvastatin bound to the HMGR active site. This 2.2 Å simulated-annealing omit map, contoured at 1 s, was calculated by omitting all atoms of the atorvastatin molecule shown, as well as protein atoms within 4.5 Å of the inhibitor. The electron density is overlaid on the final, refined model. The electron density covering atorvastatin is in green, whereas the electron density covering the protein is in blue. Carbon atoms of one of the two protein monomers are colored yellow, those of the neighboring monomer are in blue, and those of atorvastatin are in gray. In all molecules oxygen atoms are red, nitrogen atoms are blue, sulfur atoms are yellow, and the fluorine atoms are green. Fig. 4. Mode of binding of compactin (A), simvastatin (B), fluvastatin (C), cerivastatin (D), atorvastatin (E), and rosuvastatin (F) to human HMGR. Interactions between the HMG moieties of the statins and the protein are mostly ionic or polar. They are similar for all inhibitors and are indicated by the dotted lines. Numbers next to the lines indicate distances in Å (13). The rigid hydrophobic groups of the statins are situated in a shallow groove between helices La1 and La10. Additional interactions between Arg590 and the fluorophenyl group are present in the type 2 statins (C, D, E, F). Atorvastatin and rosuvastatin form a hydrogen bond between Ser565 and a carbonyl oxygen atom (atorvastatin) (E) or a sulfone oxygen atom (rosuvastatin) (F). R EPORTS 1162 11 MAY 2001 VOL 292 SCIENCE www.sciencemag.org