正在加载图片...
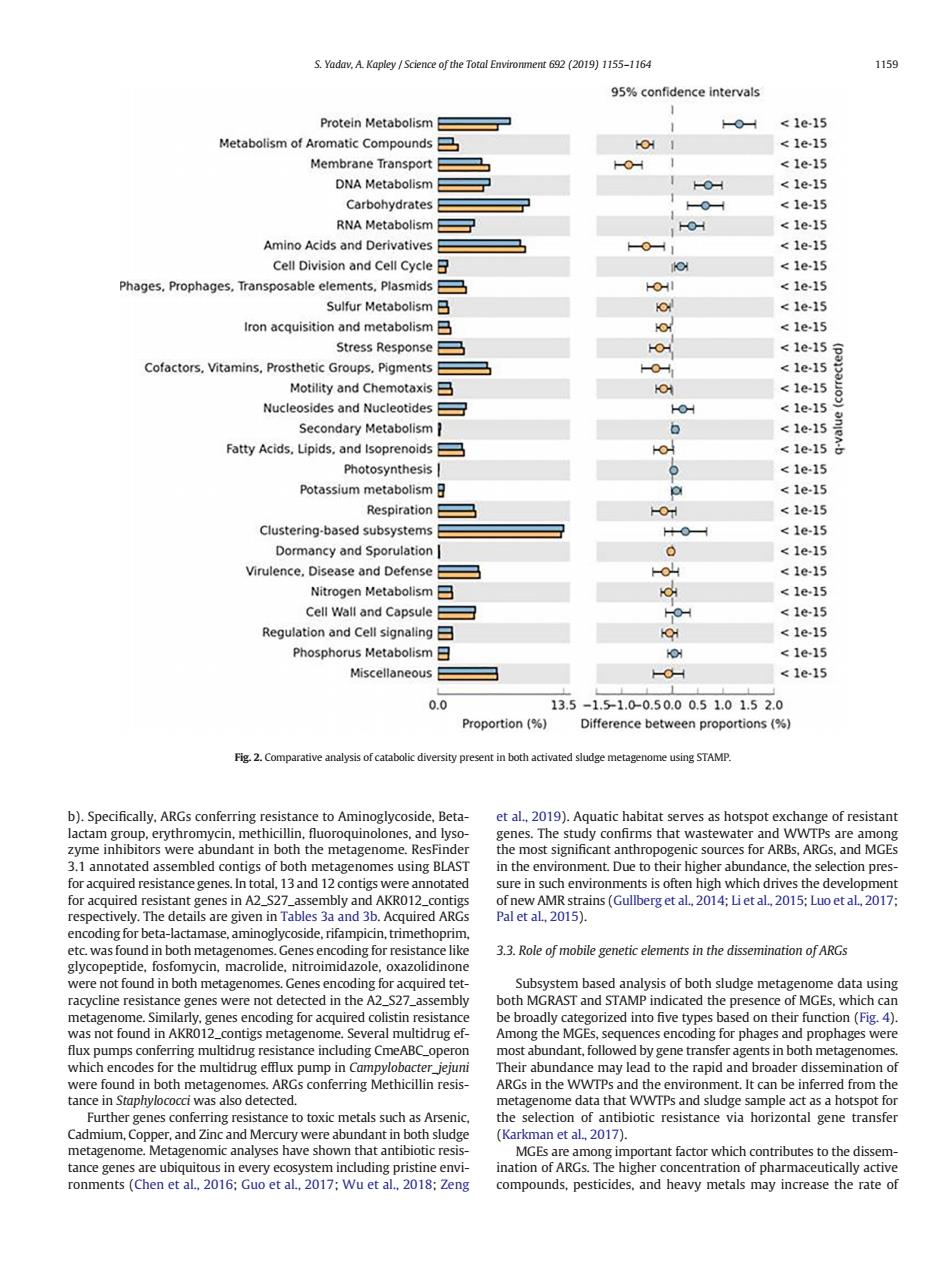
S.Yadry.A Kanley Science of the Total 692 (2019)1155-1164 159 %confidence intervals Protein Metabolism 0 1e-15 Metabolism of Ar tic Co m nds 1e1 Membrane Transpon目 11 1e15 Carbohydrates <1e-15 RNA Metabolism目 <1e-15 Amino Acids and Derivatives <1e-15 Cell Division and Cell Cycle stress Response 1e-15 Cofactors.Vitamins.Prosthetic Groups.Piaments 1e.15 Motility and Chemotaxis日 <1e-15 Nucleosides and Nucleotides -OH <1e-15 Secondary Metabolism <1e-15 Fatty Acids.Lipids.and Isoprenoids O metabolism日 Clustering-based subsystems <1e-15 0 <1e15 Virulence.Disease and Defense <1e-15 Nitrogen Metabolism目 OH <1e15 Cell Wall and Capsule日 0 <1e-15 Regulation and Cell signaling <1e-1 llaneous Proportion (% en pre e ARGs,and MGES 33.Role of mobie genetic elements in the dissemination of ARGs racyie resistance genes were not detected in theA dicated thepr ronments (Chen et al.2016:Guo et al.2017:Wu et al.2018:Zeng compounds.pesticides,and heavy metals may increase the rate ofb). Specifically, ARGs conferring resistance to Aminoglycoside, Betalactam group, erythromycin, methicillin, fluoroquinolones, and lysozyme inhibitors were abundant in both the metagenome. ResFinder 3.1 annotated assembled contigs of both metagenomes using BLAST for acquired resistance genes. In total, 13 and 12 contigs were annotated for acquired resistant genes in A2_S27_assembly and AKR012_contigs respectively. The details are given in Tables 3a and 3b. Acquired ARGs encoding for beta-lactamase, aminoglycoside, rifampicin, trimethoprim, etc. was found in both metagenomes. Genes encoding for resistance like glycopeptide, fosfomycin, macrolide, nitroimidazole, oxazolidinone were not found in both metagenomes. Genes encoding for acquired tetracycline resistance genes were not detected in the A2_S27_assembly metagenome. Similarly, genes encoding for acquired colistin resistance was not found in AKR012_contigs metagenome. Several multidrug ef- flux pumps conferring multidrug resistance including CmeABC_operon which encodes for the multidrug efflux pump in Campylobacter_jejuni were found in both metagenomes. ARGs conferring Methicillin resistance in Staphylococci was also detected. Further genes conferring resistance to toxic metals such as Arsenic, Cadmium, Copper, and Zinc and Mercury were abundant in both sludge metagenome. Metagenomic analyses have shown that antibiotic resistance genes are ubiquitous in every ecosystem including pristine environments (Chen et al., 2016; Guo et al., 2017; Wu et al., 2018; Zeng et al., 2019). Aquatic habitat serves as hotspot exchange of resistant genes. The study confirms that wastewater and WWTPs are among the most significant anthropogenic sources for ARBs, ARGs, and MGEs in the environment. Due to their higher abundance, the selection pressure in such environments is often high which drives the development of new AMR strains (Gullberg et al., 2014; Li et al., 2015; Luo et al., 2017; Pal et al., 2015). 3.3. Role of mobile genetic elements in the dissemination of ARGs Subsystem based analysis of both sludge metagenome data using both MGRAST and STAMP indicated the presence of MGEs, which can be broadly categorized into five types based on their function (Fig. 4). Among the MGEs, sequences encoding for phages and prophages were most abundant, followed by gene transfer agents in both metagenomes. Their abundance may lead to the rapid and broader dissemination of ARGs in the WWTPs and the environment. It can be inferred from the metagenome data that WWTPs and sludge sample act as a hotspot for the selection of antibiotic resistance via horizontal gene transfer (Karkman et al., 2017). MGEs are among important factor which contributes to the dissemination of ARGs. The higher concentration of pharmaceutically active compounds, pesticides, and heavy metals may increase the rate of Fig. 2. Comparative analysis of catabolic diversity present in both activated sludge metagenome using STAMP. S. Yadav, A. Kapley / Science of the Total Environment 692 (2019) 1155–1164 1159