正在加载图片...
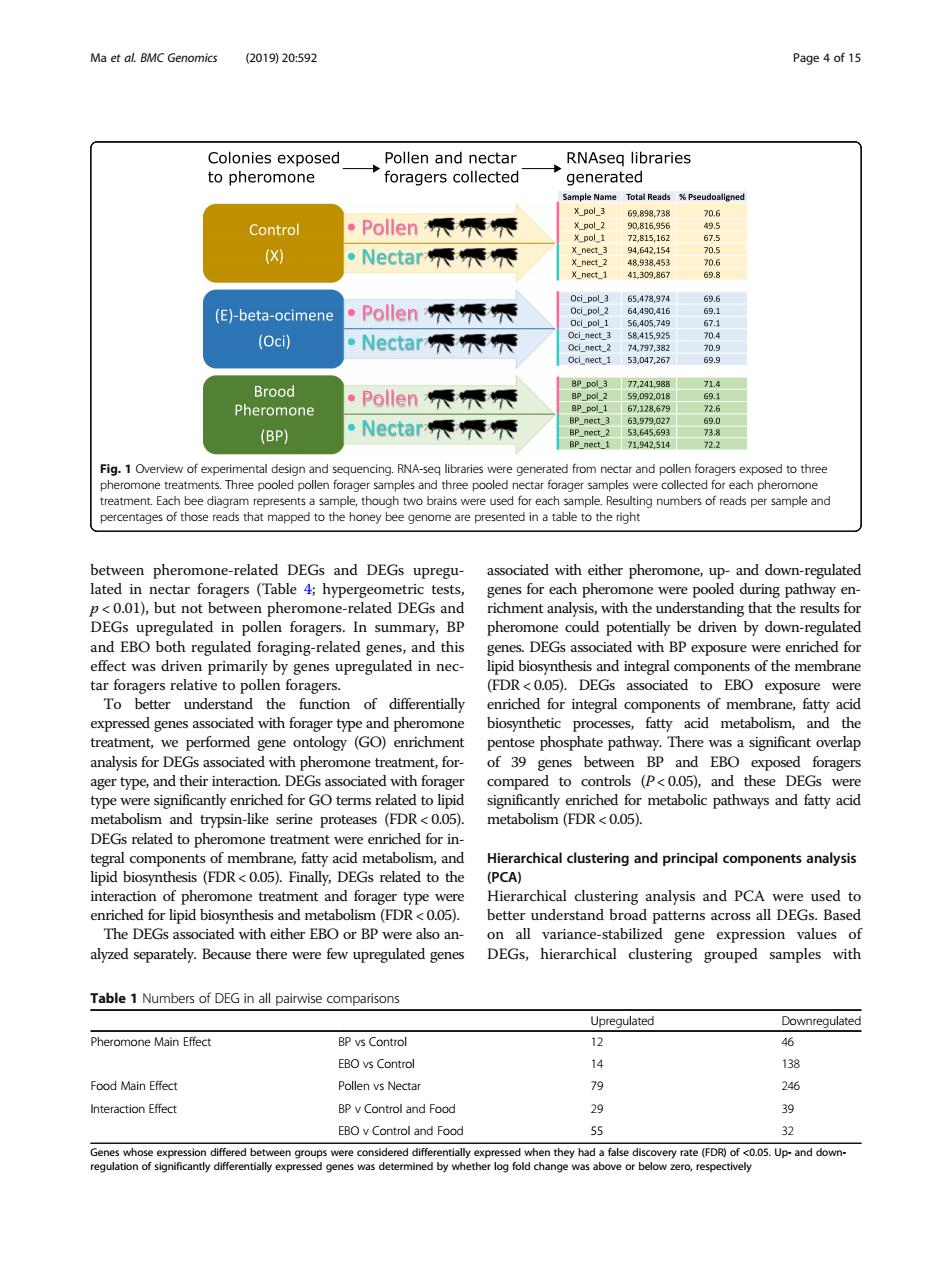
Ma et al BMC Genomics (201920592 Page 4of 15 Colonies exposed Pollen and nectar RNAseq libraries to pheromone roragers collectec generate Control ·Pollen师师 898738 X 。Nectar师辰辰 (E)-beta-ocimene ·Pollen师乘辰 (Oci】 ·Nectar辰辰 ·Pollen师师元 (BP) ·Nectar师乘元 Fig.1 Overview of e ng numbers of reads per sample and between pheromone-related DEGs and DEGs upregu lated in but not ol phe om thth ding that the result and ERO alata DEC with RD had fo effect was driven primarily by genes upregulated in nec- ipid biosynthesis and integral com onents of the membrane tar foragers relative to pollen foragers. (FDR<0 DEGs associated to EBO exposure were 10 und r integral treatment.we performed pentose phosphate pathway.There was a significant verp analysis for DEGs associated with pheromone treatment,for- of 39 genes between BP and EBO exposed foragers ager type,and their intera and these DEGs were type en tike y en DEGs related top ne treat t were enriched for in tegral components of membrane,fatty acid metabolism,and Hierarchical clustering and principal components analysis ipid biosynthe (FDR<0.05).Finally, EGs related to the PCA) chical clust ring analysis and PCA nd h The dEgs associated with either ebo or bp were also an n all variance-stabilized gene expression values of alvzed separately.Because there were few upregulated genes DEGs,hierarchical clustering grouped samples with Table 1 Numbers of DEG in all pairwise comparisons Upregulated Downregulated Pheromone Main Effect BP ys Control 12 46 EBO ys Contro 14 138 Food Main Effect Pollen ys Nectar 79 24 BP y Control and Food 30 EBOvCont and Food Up-and downbetween pheromone-related DEGs and DEGs upregulated in nectar foragers (Table 4; hypergeometric tests, p < 0.01), but not between pheromone-related DEGs and DEGs upregulated in pollen foragers. In summary, BP and EBO both regulated foraging-related genes, and this effect was driven primarily by genes upregulated in nectar foragers relative to pollen foragers. To better understand the function of differentially expressed genes associated with forager type and pheromone treatment, we performed gene ontology (GO) enrichment analysis for DEGs associated with pheromone treatment, forager type, and their interaction. DEGs associated with forager type were significantly enriched for GO terms related to lipid metabolism and trypsin-like serine proteases (FDR < 0.05). DEGs related to pheromone treatment were enriched for integral components of membrane, fatty acid metabolism, and lipid biosynthesis (FDR < 0.05). Finally, DEGs related to the interaction of pheromone treatment and forager type were enriched for lipid biosynthesis and metabolism (FDR < 0.05). The DEGs associated with either EBO or BP were also analyzed separately. Because there were few upregulated genes associated with either pheromone, up- and down-regulated genes for each pheromone were pooled during pathway enrichment analysis, with the understanding that the results for pheromone could potentially be driven by down-regulated genes. DEGs associated with BP exposure were enriched for lipid biosynthesis and integral components of the membrane (FDR < 0.05). DEGs associated to EBO exposure were enriched for integral components of membrane, fatty acid biosynthetic processes, fatty acid metabolism, and the pentose phosphate pathway. There was a significant overlap of 39 genes between BP and EBO exposed foragers compared to controls (P < 0.05), and these DEGs were significantly enriched for metabolic pathways and fatty acid metabolism (FDR < 0.05). Hierarchical clustering and principal components analysis (PCA) Hierarchical clustering analysis and PCA were used to better understand broad patterns across all DEGs. Based on all variance-stabilized gene expression values of DEGs, hierarchical clustering grouped samples with Fig. 1 Overview of experimental design and sequencing. RNA-seq libraries were generated from nectar and pollen foragers exposed to three pheromone treatments. Three pooled pollen forager samples and three pooled nectar forager samples were collected for each pheromone treatment. Each bee diagram represents a sample, though two brains were used for each sample. Resulting numbers of reads per sample and percentages of those reads that mapped to the honey bee genome are presented in a table to the right Table 1 Numbers of DEG in all pairwise comparisons Upregulated Downregulated Pheromone Main Effect BP vs Control 12 46 EBO vs Control 14 138 Food Main Effect Pollen vs Nectar 79 246 Interaction Effect BP v Control and Food 29 39 EBO v Control and Food 55 32 Genes whose expression differed between groups were considered differentially expressed when they had a false discovery rate (FDR) of <0.05. Up- and downregulation of significantly differentially expressed genes was determined by whether log fold change was above or below zero, respectively Ma et al. BMC Genomics (2019) 20:592 Page 4 of 15