正在加载图片...
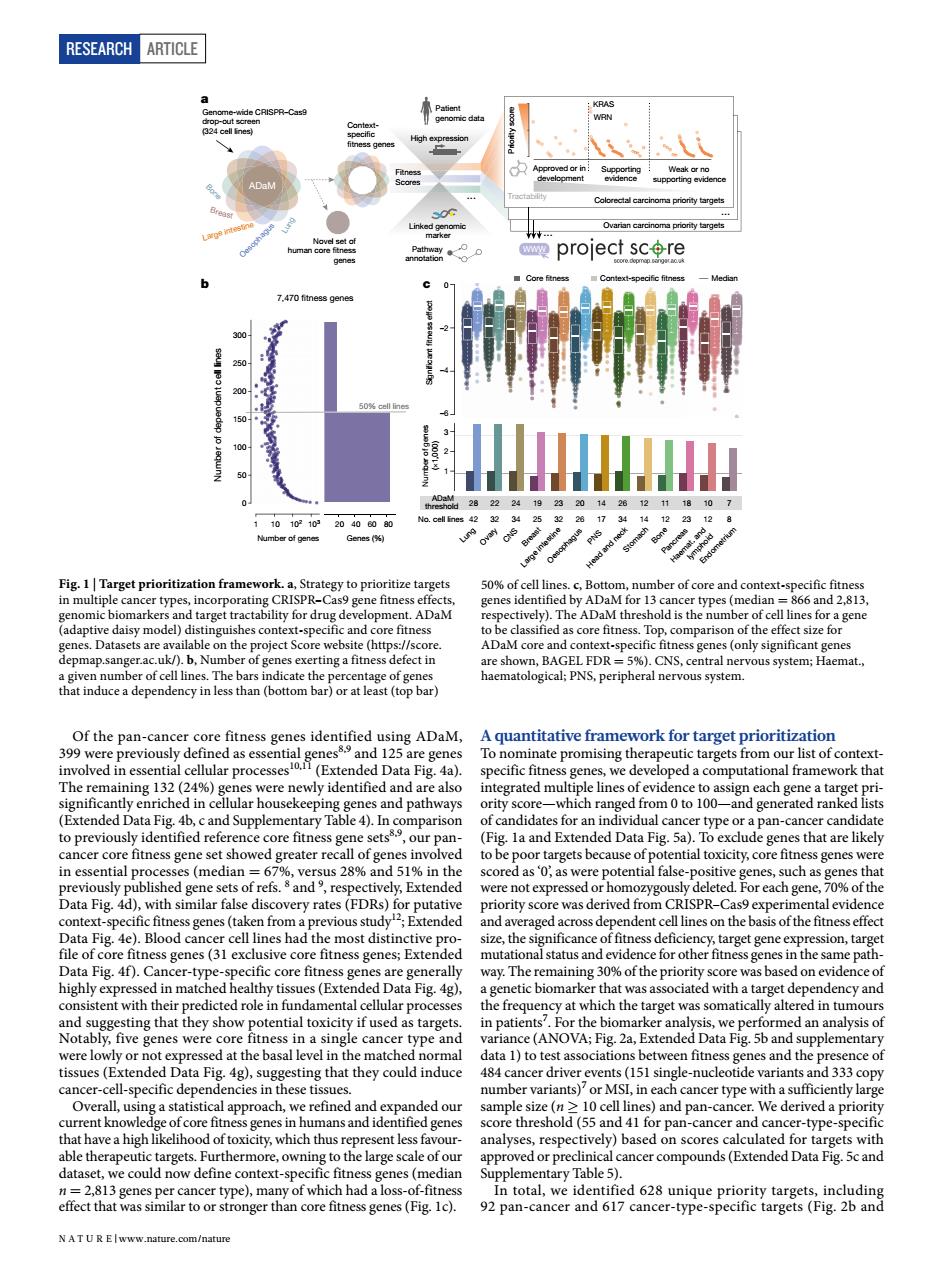
RESEARCH ARTICLE project scre hD)CNc ADaM core and aaakarcaetpnitaionc y path )T起a enes that are likely nut stent 盈球co水map side varian o9aama 10 cell lines)and pa We deriveda pr nt less fa cancer compounds (Extended Data Fig.5c andRESEARCH Article Of the pan-cancer core fitness genes identified using ADaM, 399 were previously defined as essential genes8,9 and 125 are genes involved in essential cellular processes10,11 (Extended Data Fig. 4a). The remaining 132 (24%) genes were newly identified and are also significantly enriched in cellular housekeeping genes and pathways (Extended Data Fig. 4b, c and Supplementary Table 4). In comparison to previously identified reference core fitness gene sets8,9 , our pancancer core fitness gene set showed greater recall of genes involved in essential processes (median = 67%, versus 28% and 51% in the previously published gene sets of refs. 8 and 9 , respectively, Extended Data Fig. 4d), with similar false discovery rates (FDRs) for putative context-specific fitness genes (taken from a previous study12; Extended Data Fig. 4e). Blood cancer cell lines had the most distinctive profile of core fitness genes (31 exclusive core fitness genes; Extended Data Fig. 4f). Cancer-type-specific core fitness genes are generally highly expressed in matched healthy tissues (Extended Data Fig. 4g), consistent with their predicted role in fundamental cellular processes and suggesting that they show potential toxicity if used as targets. Notably, five genes were core fitness in a single cancer type and were lowly or not expressed at the basal level in the matched normal tissues (Extended Data Fig. 4g), suggesting that they could induce cancer-cell-specific dependencies in these tissues. Overall, using a statistical approach, we refined and expanded our current knowledge of core fitness genes in humans and identified genes that have a high likelihood of toxicity, which thus represent less favourable therapeutic targets. Furthermore, owning to the large scale of our dataset, we could now define context-specific fitness genes (median n = 2,813 genes per cancer type), many of which had a loss-of-fitness effect that was similar to or stronger than core fitness genes (Fig. 1c). A quantitative framework for target prioritization To nominate promising therapeutic targets from our list of contextspecific fitness genes, we developed a computational framework that integrated multiple lines of evidence to assign each gene a target priority score—which ranged from 0 to 100—and generated ranked lists of candidates for an individual cancer type or a pan-cancer candidate (Fig. 1a and Extended Data Fig. 5a). To exclude genes that are likely to be poor targets because of potential toxicity, core fitness genes were scored as ‘0’, as were potential false-positive genes, such as genes that were not expressed or homozygously deleted. For each gene, 70% of the priority score was derived from CRISPR–Cas9 experimental evidence and averaged across dependent cell lines on the basis of the fitness effect size, the significance of fitness deficiency, target gene expression, target mutational status and evidence for other fitness genes in the same pathway. The remaining 30% of the priority score was based on evidence of a genetic biomarker that was associated with a target dependency and the frequency at which the target was somatically altered in tumours in patients7 . For the biomarker analysis, we performed an analysis of variance (ANOVA; Fig. 2a, Extended Data Fig. 5b and supplementary data 1) to test associations between fitness genes and the presence of 484 cancer driver events (151 single-nucleotide variants and 333 copy number variants)7 or MSI, in each cancer type with a sufficiently large sample size (n ≥ 10 cell lines) and pan-cancer. We derived a priority score threshold (55 and 41 for pan-cancer and cancer-type-specific analyses, respectively) based on scores calculated for targets with approved or preclinical cancer compounds (Extended Data Fig. 5c and Supplementary Table 5). In total, we identified 628 unique priority targets, including 92 pan-cancer and 617 cancer-type-specific targets (Fig. 2b and Ovarian carcinoma priority targets Novel set of human core tness genes Oesophagus Large intestine Breast Bone ADaM Patient genomic data Priority score KRAS WRN Colorectal carcinoma priority targets Approved or in development Supporting evidence Weak or no supporting evidence Linked genomic marker High expression ... Tractability Fitness Scores Contextspeci c tness genes Pathway annotation Genome-wide CRISPR–Cas9 drop-out screen (324 cell lines) Lung ... ... 42 32 34 25 32 26 17 34 14 12 23 12 8 28 22 24 19 23 20 14 26 12 11 18 10 7 No. cell lines ADaM threshold Number of genes (×1,000) 1 2 3 Signi cant tness effect –2 0 –4 –6 Core tness Context-speci c tness Median b a c 7,470 tness genes Number of dependent cell lines 0 50 100 150 200 250 300 1 10 102 103 Number of genes 40 60 80 Genes (%) 20 Lung Ovary CNS Breast Large intestine Oesophagus PNS Head and neck Stomach Bone Haemat. and lymphoid Pancreas Endometrium 50% cell lines WWW Fig. 1 | Target prioritization framework. a, Strategy to prioritize targets in multiple cancer types, incorporating CRISPR–Cas9 gene fitness effects, genomic biomarkers and target tractability for drug development. ADaM (adaptive daisy model) distinguishes context-specific and core fitness genes. Datasets are available on the project Score website (https://score. depmap.sanger.ac.uk/). b, Number of genes exerting a fitness defect in a given number of cell lines. The bars indicate the percentage of genes that induce a dependency in less than (bottom bar) or at least (top bar) 50% of cell lines. c, Bottom, number of core and context-specific fitness genes identified by ADaM for 13 cancer types (median = 866 and 2,813, respectively). The ADaM threshold is the number of cell lines for a gene to be classified as core fitness. Top, comparison of the effect size for ADaM core and context-specific fitness genes (only significant genes are shown, BAGEL FDR = 5%). CNS, central nervous system; Haemat., haematological; PNS, peripheral nervous system. N A t U r e | www.nature.com/nature