正在加载图片...
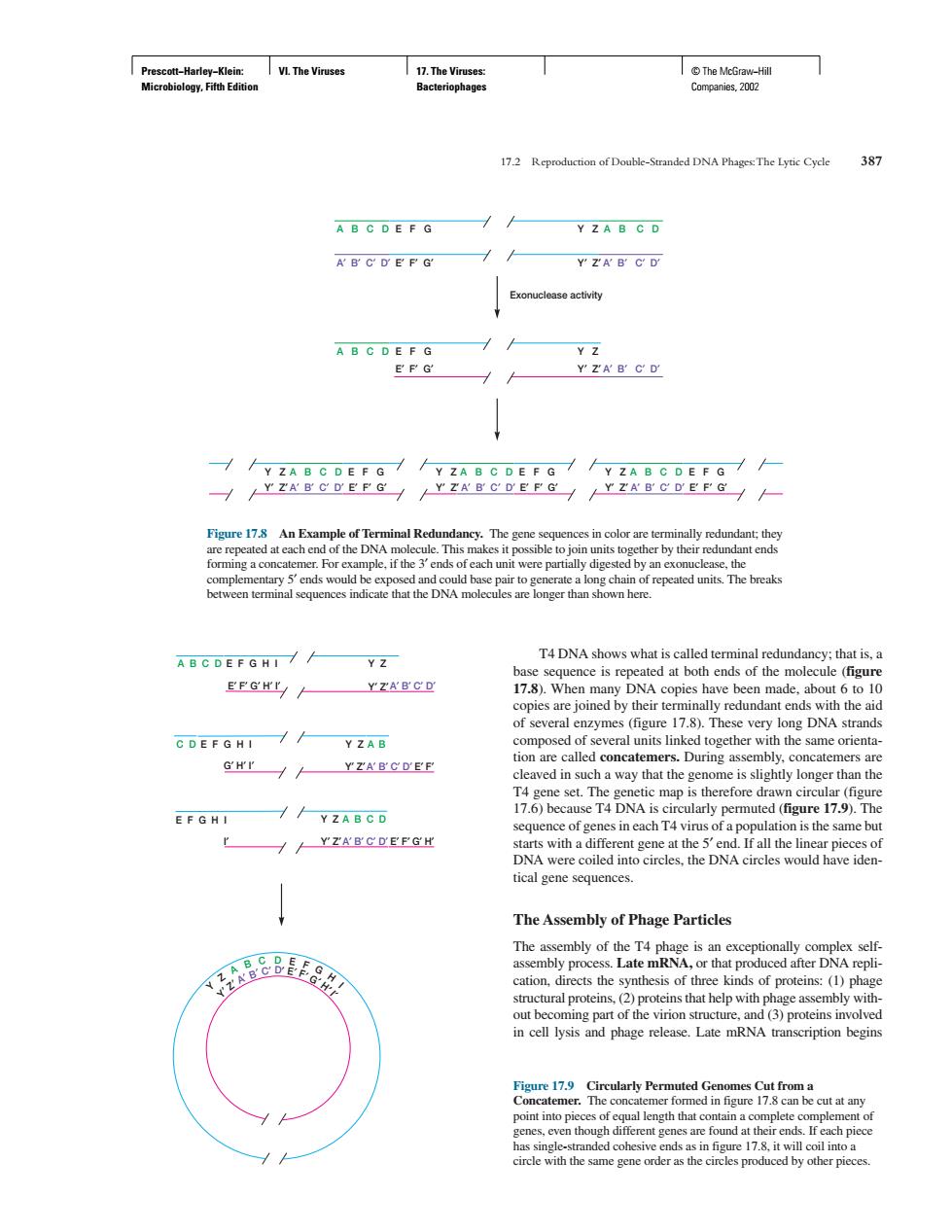
I VL The Viruse we 172 Reproduction of Double-Standed DNA The Lytic Cyde 31 A B C D E F G Y ZA B G D A B C D E P G Y ZA B C D Exonuclease activity A B C D E F G YZAB C D n each u AB C DEFG HI// T4 DNA shows what is called ter Y Z of several enzymes (figure 17.8).These very long DNA strands C DEFGHI / Y ZAB GH'T H YZABCDEF cleaved in such a way that the genome is slightly longer than the //YZABCD r /YZARCDEEGH is the same bu DNA were DNA circles would have iden tical gene sequences. The Assembly of Phage Particles m) begin ut at an gointin Prescott−Harley−Klein: Microbiology, Fifth Edition VI. The Viruses 17. The Viruses: Bacteriophages © The McGraw−Hill Companies, 2002 T4 DNA shows what is called terminal redundancy; that is, a base sequence is repeated at both ends of the molecule (figure 17.8). When many DNA copies have been made, about 6 to 10 copies are joined by their terminally redundant ends with the aid of several enzymes (figure 17.8). These very long DNA strands composed of several units linked together with the same orientation are called concatemers. During assembly, concatemers are cleaved in such a way that the genome is slightly longer than the T4 gene set. The genetic map is therefore drawn circular (figure 17.6) because T4 DNA is circularly permuted (figure 17.9). The sequence of genes in each T4 virus of a population is the same but starts with a different gene at the 5′ end. If all the linear pieces of DNA were coiled into circles, the DNA circles would have identical gene sequences. The Assembly of Phage Particles The assembly of the T4 phage is an exceptionally complex selfassembly process. Late mRNA, or that produced after DNA replication, directs the synthesis of three kinds of proteins: (1) phage structural proteins, (2) proteins that help with phage assembly without becoming part of the virion structure, and (3) proteins involved in cell lysis and phage release. Late mRNA transcription begins 17.2 Reproduction of Double-Stranded DNA Phages:The Lytic Cycle 387 Y′ Z′ A′ B′ C′ D′ E′ F′ G′ Y′ Z′ A′ B′ C′ D′ E′ F′ G′ Y′ Z′ A′ B′ C′ D′ E′ F′ G′ A B C D A′ B′ C′ D′ A B C D E F G Y Z A′ B′ C′ D′ E′ F′ G′ Y′ Z′ Exonuclease activity E F G Y Z E′ F′ G′ Y′ Z′ A B C D A′ B′ C′ D′ Y Z A B C D E F G Y Z A B C D E F G Y Z A B C D E F G Figure 17.8 An Example of Terminal Redundancy. The gene sequences in color are terminally redundant; they are repeated at each end of the DNA molecule. This makes it possible to join units together by their redundant ends forming a concatemer. For example, if the 3′ ends of each unit were partially digested by an exonuclease, the complementary 5′ ends would be exposed and could base pair to generate a long chain of repeated units. The breaks between terminal sequences indicate that the DNA molecules are longer than shown here. A′ B′ C′ D′ E F G H I A′ B′ C′ D′ A B C D E F G H I Y Z E′ F′ G′ H′ I′ Y′ Z′ C D E F G H I Y Z A B G′ H′ I′ Y′ Z′ A′ B′ C′ D′ Y Z I′ Y′ Z′ A B C D E′ F′ E′ F′ G′ H′ Y Z A B C D E F G H I Y′ Z′ A′ B′ C′ D′ E′ F′ G′ H′ I′ Figure 17.9 Circularly Permuted Genomes Cut from a Concatemer. The concatemer formed in figure 17.8 can be cut at any point into pieces of equal length that contain a complete complement of genes, even though different genes are found at their ends. If each piece has single-stranded cohesive ends as in figure 17.8, it will coil into a circle with the same gene order as the circles produced by other pieces