正在加载图片...
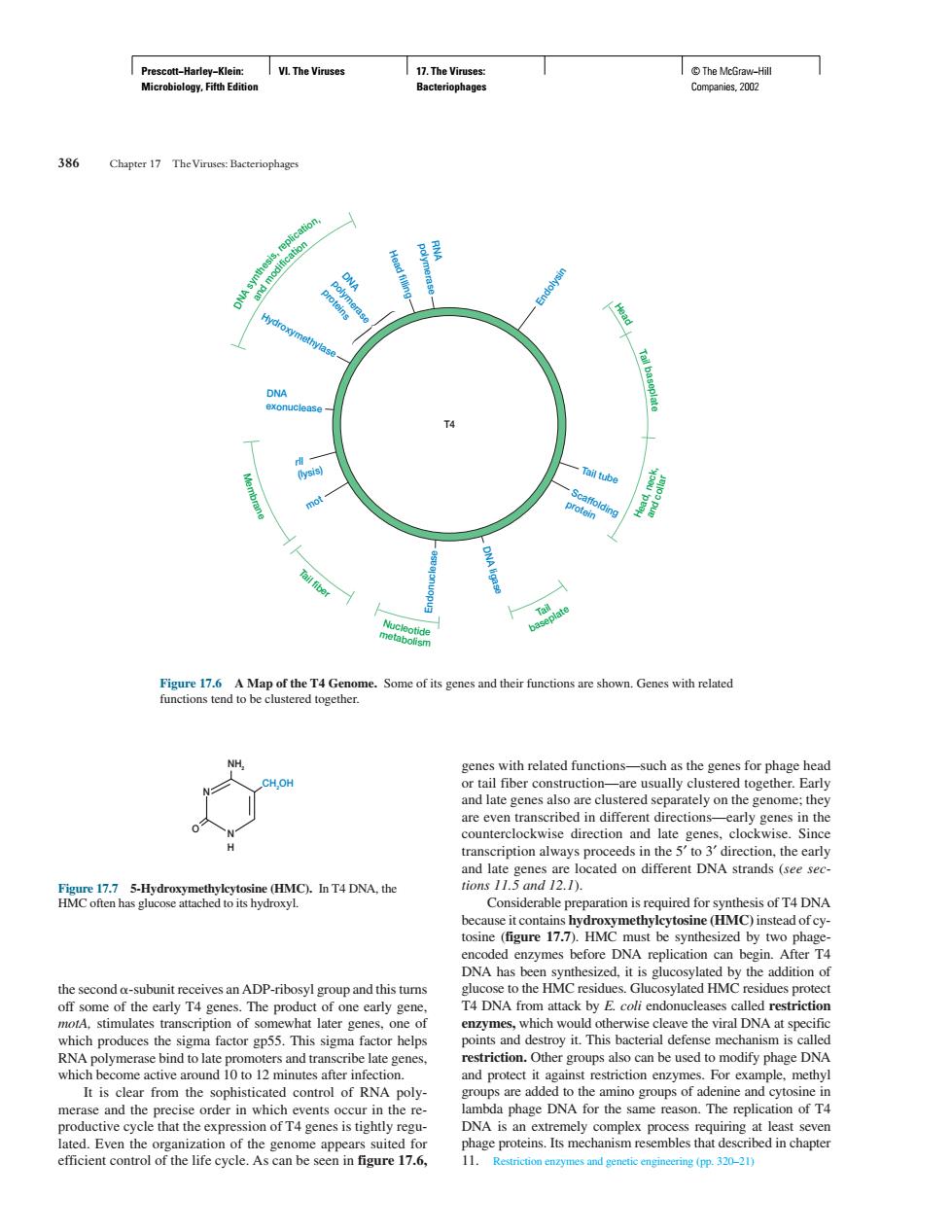
ww The Viruses:Bacteriophoge Some of its ec s and their func shown Genes with related or tail fiber Ce空IO.hT4D, nlate enes on differe DNA strands ation isrequred for synthesis of T4 DNA lcytosine(HMC)instead of ey- DNA has ben synth zed.it is glucosy stim restriction other s s also can be used to modify phage DNA amla phae DNA for The of T efficient control of the life cycle.Ascan be sen.6. 11. (DP:320-21Prescott−Harley−Klein: Microbiology, Fifth Edition VI. The Viruses 17. The Viruses: Bacteriophages © The McGraw−Hill Companies, 2002 the second -subunit receives an ADP-ribosyl group and this turns off some of the early T4 genes. The product of one early gene, motA, stimulates transcription of somewhat later genes, one of which produces the sigma factor gp55. This sigma factor helps RNA polymerase bind to late promoters and transcribe late genes, which become active around 10 to 12 minutes after infection. It is clear from the sophisticated control of RNA polymerase and the precise order in which events occur in the reproductive cycle that the expression of T4 genes is tightly regulated. Even the organization of the genome appears suited for efficient control of the life cycle. As can be seen in figure 17.6, genes with related functions—such as the genes for phage head or tail fiber construction—are usually clustered together. Early and late genes also are clustered separately on the genome; they are even transcribed in different directions—early genes in the counterclockwise direction and late genes, clockwise. Since transcription always proceeds in the 5′ to 3′ direction, the early and late genes are located on different DNA strands (see sections 11.5 and 12.1). Considerable preparation is required for synthesis of T4 DNA because it contains hydroxymethylcytosine (HMC) instead of cytosine (figure 17.7). HMC must be synthesized by two phageencoded enzymes before DNA replication can begin. After T4 DNA has been synthesized, it is glucosylated by the addition of glucose to the HMC residues. Glucosylated HMC residues protect T4 DNA from attack by E. coli endonucleases called restriction enzymes, which would otherwise cleave the viral DNA at specific points and destroy it. This bacterial defense mechanism is called restriction. Other groups also can be used to modify phage DNA and protect it against restriction enzymes. For example, methyl groups are added to the amino groups of adenine and cytosine in lambda phage DNA for the same reason. The replication of T4 DNA is an extremely complex process requiring at least seven phage proteins. Its mechanism resembles that described in chapter 11. Restriction enzymes and genetic engineering (pp. 320–21) 386 Chapter 17 The Viruses: Bacteriophages Hydroxymethylase Head filling polymerase RNA Head, neck, and modification Endolysin Tail tube Scaffolding protein DNA ligase Endonuclease rll (lysis) mot Head Tail baseplate and collar m Nucleotide etabolism Tail fiber Membrane T4 DNA synthesis, replication, DNA exonuclease DNA polymerase proteins Tail baseplate Figure 17.6 A Map of the T4 Genome. Some of its genes and their functions are shown. Genes with related functions tend to be clustered together. CH2 OH NH2 O N H N Figure 17.7 5-Hydroxymethylcytosine (HMC). In T4 DNA, the HMC often has glucose attached to its hydroxyl