正在加载图片...
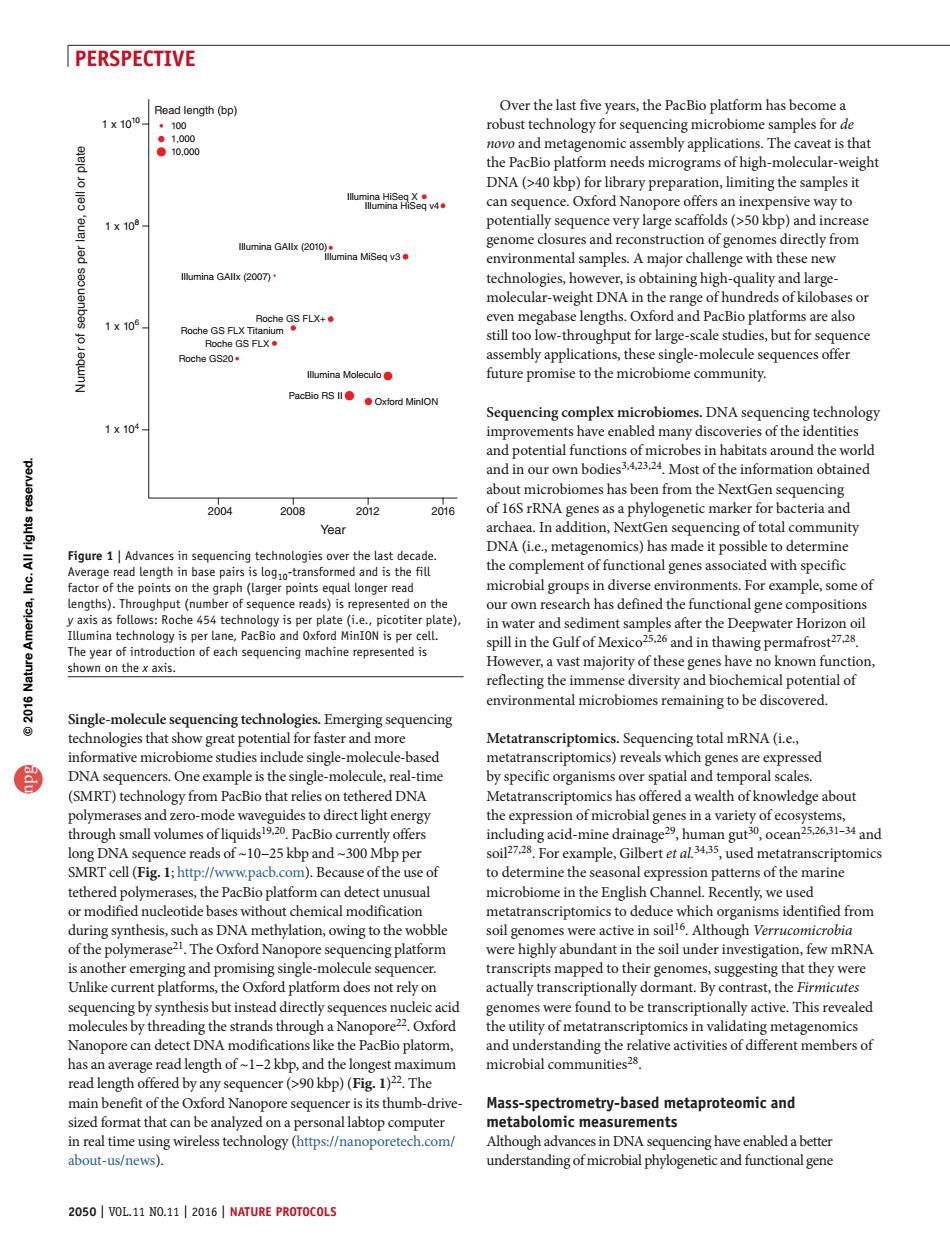
PERSPECTIVE 1x100 length (bp) Over the last five years,the PacBio platform has becomea robust tec the PacBio platform needs micrograms ofigh-molecua-weigh DNA(>40 kbp)for library preparation,limiting the samples it nee 1x10 and increas environmental samples.A maior challenge with these new technologies,however,is obtaining high-quality and large molecular-weight D A in the range of hundreds ofkilob 501 and oche G520 gle-molecule sequ nces offer future promise to the microbiome community. Sequencing complex mi DNA sequ ogy 1x104 functions ofmi bes in habitats around the world and in our own bodies3 4 3.24.Most of the information obtained about microbiomes has b n from the NextGen sequencing 2004 2008 2012 2016 a phylogenetic ma teria an Year t omics)has made it p the complement of functional genes associated with specific agfhepoieonthe aph (a microbial groups in diverse environments.For example,some of s repre our own re has detine the rur tonal gene c ncin pill in the Gulf of h nafrost27.2s shown on the x axis However.a vast majority of these genes have o known function reflecting the immense diversity and biochemical potential of environmental microbiomes remaining to be discovered sequencing technolo onies that s tial for fa Metatrans ng total mrNa (ie. informative microbiome studies indude single-molecule-based metatranscriptomics)reveals which genes are exp sed DNA sequencers.One example is the single-molecule,real-time by specific organisms over spatial and temporal scales. (SMRT)technology from PacBio that relies on tethered DNA Metatranscriptomics ha ffered a wealth wledge abou polym na varie DNA sequence reads of -10-25 kbp and-300 Mbp per sFor exampe Gilbert mctatranscriptomics SMRT cell (Fig1:http://www.pacb.com).Because ofthe use of to determine the seasonal expression patterns of the marine microbiome in the English Chann Recently,we used m uch as DNA platform were highly abundant in the soil under investigation.few mRNA transcripts mapped to their genomes,suggesting g that they were I pla orm doe s not rely on ctually trans nally dormant.By contrast,the F ly active.This rev Nanopore can detect DNA modifications like the PacBio platorm has an average read length of~1-2 kbp,and the longes microbial communities?8 ngth ofle rd Nai ore sequenc its thumb- etry-based about-us/news) understanding of microbialphylogenetic and functional gene 2050 VOL.11 NO.NATURE PROTOCOLS 2050 | VOL.11 NO.11 | 2016 | NATURE PROTOCOLS PERSPECTIVE Over the last five years, the PacBio platform has become a robust technology for sequencing microbiome samples for de novo and metagenomic assembly applications. The caveat is that the PacBio platform needs micrograms of high-molecular-weight DNA (>40 kbp) for library preparation, limiting the samples it can sequence. Oxford Nanopore offers an inexpensive way to potentially sequence very large scaffolds (>50 kbp) and increase genome closures and reconstruction of genomes directly from environmental samples. A major challenge with these new technologies, however, is obtaining high-quality and largemolecular-weight DNA in the range of hundreds of kilobases or even megabase lengths. Oxford and PacBio platforms are also still too low-throughput for large-scale studies, but for sequence assembly applications, these single-molecule sequences offer future promise to the microbiome community. Sequencing complex microbiomes. DNA sequencing technology improvements have enabled many discoveries of the identities and potential functions of microbes in habitats around the world and in our own bodies3,4,23,24. Most of the information obtained about microbiomes has been from the NextGen sequencing of 16S rRNA genes as a phylogenetic marker for bacteria and archaea. In addition, NextGen sequencing of total community DNA (i.e., metagenomics) has made it possible to determine the complement of functional genes associated with specific microbial groups in diverse environments. For example, some of our own research has defined the functional gene compositions in water and sediment samples after the Deepwater Horizon oil spill in the Gulf of Mexico25,26 and in thawing permafrost27,28. However, a vast majority of these genes have no known function, reflecting the immense diversity and biochemical potential of environmental microbiomes remaining to be discovered. Metatranscriptomics. Sequencing total mRNA (i.e., metatranscriptomics) reveals which genes are expressed by specific organisms over spatial and temporal scales. Metatranscriptomics has offered a wealth of knowledge about the expression of microbial genes in a variety of ecosystems, including acid-mine drainage29, human gut30, ocean25,26,31–34 and soil27,28. For example, Gilbert et al.34,35, used metatranscriptomics to determine the seasonal expression patterns of the marine microbiome in the English Channel. Recently, we used metatranscriptomics to deduce which organisms identified from soil genomes were active in soil16. Although Verrucomicrobia were highly abundant in the soil under investigation, few mRNA transcripts mapped to their genomes, suggesting that they were actually transcriptionally dormant. By contrast, the Firmicutes genomes were found to be transcriptionally active. This revealed the utility of metatranscriptomics in validating metagenomics and understanding the relative activities of different members of microbial communities28. Mass-spectrometry-based metaproteomic and metabolomic measurements Although advances in DNA sequencing have enabled a better understanding of microbial phylogenetic and functional gene Single-molecule sequencing technologies. Emerging sequencing technologies that show great potential for faster and more informative microbiome studies include single-molecule-based DNA sequencers. One example is the single-molecule, real-time (SMRT) technology from PacBio that relies on tethered DNA polymerases and zero-mode waveguides to direct light energy through small volumes of liquids19,20. PacBio currently offers long DNA sequence reads of ~10-25 kbp and ~300 Mbp per SMRT cell (Fig. 1; http://www.pacb.com). Because of the use of tethered polymerases, the PacBio platform can detect unusual or modified nucleotide bases without chemical modification during synthesis, such as DNA methylation, owing to the wobble of the polymerase21. The Oxford Nanopore sequencing platform is another emerging and promising single-molecule sequencer. Unlike current platforms, the Oxford platform does not rely on sequencing by synthesis but instead directly sequences nucleic acid molecules by threading the strands through a Nanopore22. Oxford Nanopore can detect DNA modifications like the PacBio platorm, has an average read length of ~1-2 kbp, and the longest maximum read length offered by any sequencer (>90 kbp) (Fig. 1)22. The main benefit of the Oxford Nanopore sequencer is its thumb-drivesized format that can be analyzed on a personal labtop computer in real time using wireless technology (https://nanoporetech.com/ about-us/news). Read length (bp) 100 1,000 10,000 Illumina HiSeq X Illumina GAIIx (2010) Illumina GAIIx (2007) Illumina MiSeq v3 Illumina Moleculo PacBio RS II Oxford MinION Illumina HiSeq v4 Roche GS FLX+ Roche GS FLX Titanium Roche GS FLX Roche GS20 1 x 1010 1 x 108 1 x 106 1 x 104 Number of sequences per lane, cell or plate Year 2004 2008 2012 2016 Figure 1 | Advances in sequencing technologies over the last decade. Average read length in base pairs is log10-transformed and is the fill factor of the points on the graph (larger points equal longer read lengths). Throughput (number of sequence reads) is represented on the y axis as follows: Roche 454 technology is per plate (i.e., picotiter plate), Illumina technology is per lane, PacBio and Oxford MinION is per cell. The year of introduction of each sequencing machine represented is shown on the x axis. npg © 2016 Nature America, Inc. All rights reserved