正在加载图片...
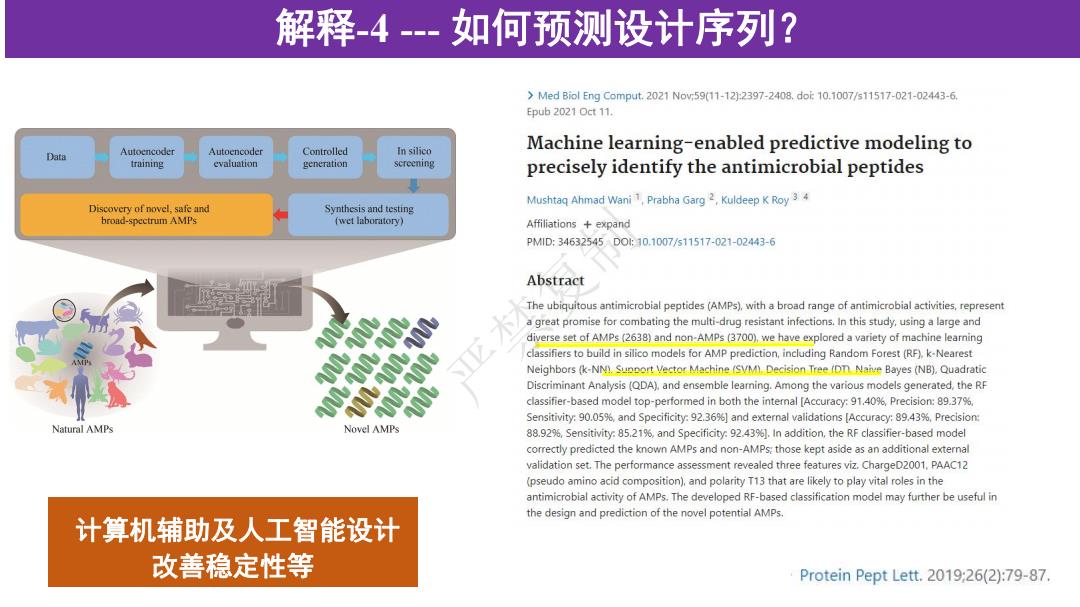
解释-4--如何预测设计序列? >Med Biol Eng Comput..2021Now59(11-122397-2408.dot10.1007/s11517-021-02443-6. Epub 2021 Oct 11. Data Autoencoder Autoencoder Controlled In silico Machine learning-enabled predictive modeling to training evaluation generation screening precisely identify the antimicrobial peptides Mushtaq Ahmad Wani1,Prabha Garg 2,Kuldeep K Roy34 Discovery of novel,safe and Synthesis and testing broad-spectrum AMPs (wet laboratory) Affiliations expand PMlD:34632545D0:10.1007/s11517-021-02443-6 Abstract The ubiquitous antimicrobial peptides(AMPs),with a broad range of antimicrobial activities,represent a great promise for combating the multi-drug resistant infections.In this study,using a large and diverse set of AMPs (2638)and non-AMPs(3700),we have explored a variety of machine learning classifiers to build in silico models for AMP prediction,including Random Forest(RF).k-Nearest Neighbors(k-NN).Support Vector Machine (SVM)Decision Tree (OT Naive Bayes (NB).Quadratic Discriminant Analysis (QDA),and ensemble leaming.Among the various models generated,the RF classifier-based model top-performed in both the intemal [Accuracy:91.40%,Precision:89.37%. Sensitivity:90.05%and Specificity:92.36%]and external validations [Accuracy:89.43%,Precision Natural AMPs Novel AMPs 88.929,Sensitivity:85.21%,and Specificity:92.43%].In addition,the RF classifier-based model correctly predicted the known AMPs and non-AMPs:those kept aside as an additional external validation set.The performance assessment revealed three features viz.ChargeD2001,PAAC12 (pseudo amino acid composition).and polarity T13 that are likely to play vital roles in the antimicrobial activity of AMPs.The developed RF-based classification model may further be useful in the design and prediction of the novel potential AMPs. 计算机辅助及人工智能设计 改善稳定性等 Protein Pept Lett.2019:26(2):79-87.解释-4 --- 如何预测设计序列? 计算机辅助及人工智能设计 改善稳定性等 严禁复制