正在加载图片...
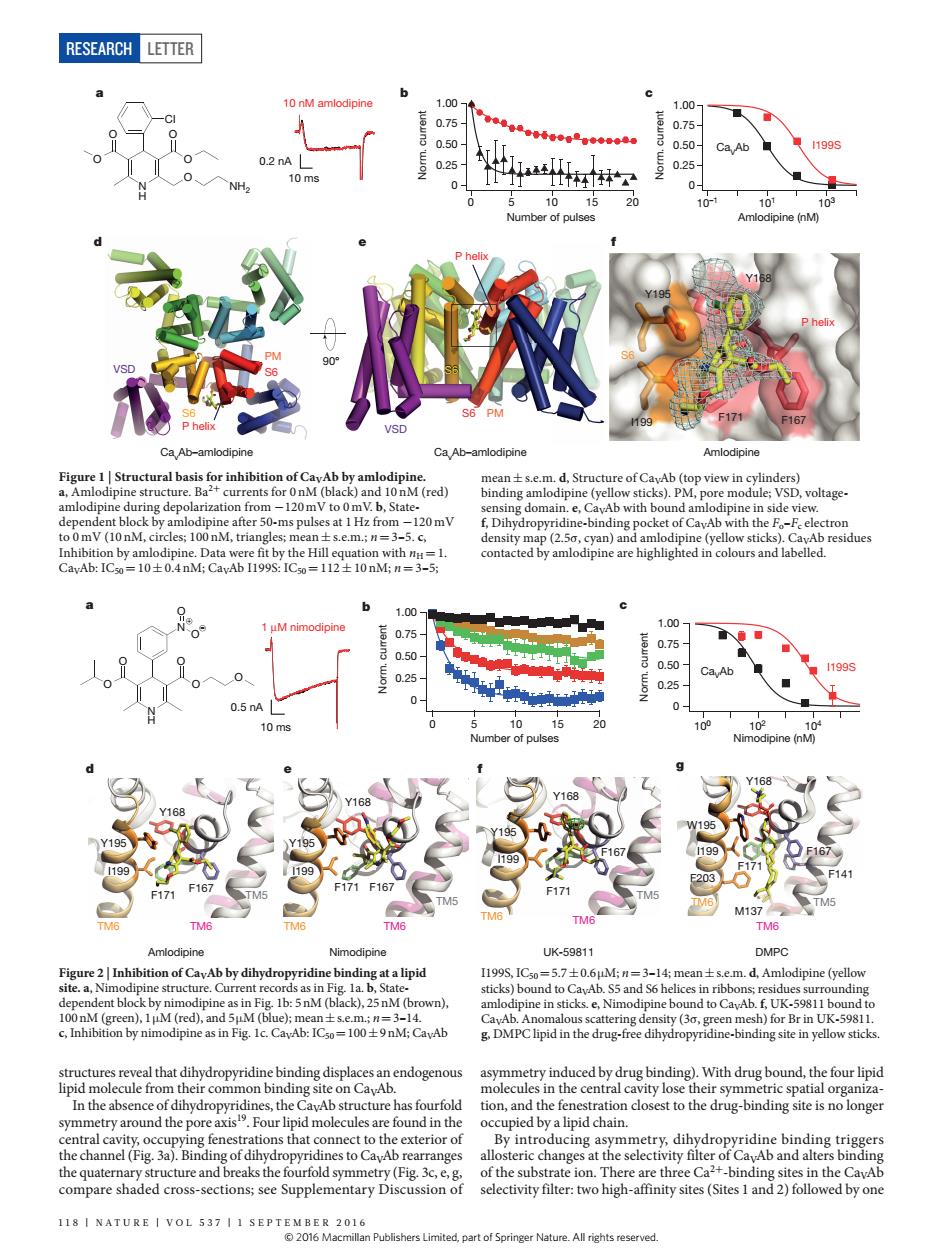
RESEARCH LETTER 织 of(red) pine in s 120mV Ca.Ab 1199 0.25 10 199 199 ● cture.Cur 57士06 g fenes ations that connect to the exterior of oducing aymmetry.dibyrn sectivyewo higyite(Sitesad)followed byone 11NATURE I VOL 537 I1 SEPTEOMmn Publishers er Nature.All tights reserved RESEARCH Letter 118 | NA T U RE | VOL 537 | 1 september 2016 structures reveal that dihydropyridine binding displaces an endogenous lipid molecule from their common binding site on CaVAb. In the absence of dihydropyridines, the CaVAb structure has fourfold symmetry around the pore axis19. Four lipid molecules are found in the central cavity, occupying fenestrations that connect to the exterior of the channel (Fig. 3a). Binding of dihydropyridines to CaVAb rearranges the quaternary structure and breaks the fourfold symmetry (Fig. 3c, e, g, compare shaded cross-sections; see Supplementary Discussion of asymmetry induced by drug binding). With drug bound, the four lipid molecules in the central cavity lose their symmetric spatial organization, and the fenestration closest to the drug-binding site is no longer occupied by a lipid chain. By introducing asymmetry, dihydropyridine binding triggers allosteric changes at the selectivity filter of CaVAb and alters binding of the substrate ion. There are three Ca2+-binding sites in the CaVAb selectivity filter: two high-affinity sites (Sites 1 and 2) followed by one Figure 1 | Structural basis for inhibition of CaVAb by amlodipine. a, Amlodipine structure. Ba2+ currents for 0 nM (black) and 10 nM (red) amlodipine during depolarization from −120 mV to 0 mV. b, Statedependent block by amlodipine after 50-ms pulses at 1Hz from −120 mV to 0 mV (10 nM, circles; 100 nM, triangles; mean±s.e.m.; n=3–5. c, Inhibition by amlodipine. Data were fit by the Hill equation with nH=1. CaVAb: IC50=10±0.4 nM; CaVAb I199S: IC50=112±10 nM; n=3–5; mean±s.e.m. d, Structure of CaVAb (top view in cylinders) binding amlodipine (yellow sticks). PM, pore module; VSD, voltagesensing domain. e, CaVAb with bound amlodipine in side view. f, Dihydropyridine-binding pocket of CaVAb with the Fo–Fc electron density map (2.5σ, cyan) and amlodipine (yellow sticks). CaVAb residues contacted by amlodipine are highlighted in colours and labelled. a e VSD PM 90° f b S6 S6 Amlodipine Y195 Y168 I199 F171 F167 P helix Cl O O O O O N H NH2 c d CavAb–amlodipine CavAb–amlodipine S6 P helix VSD PM S6 P helix S6 10 ms 0.2 nA 10 nM amlodipine 10–1 101 103 Amlodipine (nM) I199S 0.25 0.50 0.75 1.00 0 Norm. current 0.25 0.50 0.75 1.00 0 Norm. current 0 5 10 15 20 Number of pulses CaVAb Figure 2 | Inhibition of CaVAb by dihydropyridine binding at a lipid site. a, Nimodipine structure. Current records as in Fig. 1a. b, Statedependent block by nimodipine as in Fig. 1b: 5nM (black), 25nM (brown), 100nM (green), 1μM (red), and 5μM (blue); mean±s.e.m.; n=3–14. c, Inhibition by nimodipine as in Fig. 1c. CaVAb: IC50=100±9nM; CaVAb I199S, IC50=5.7±0.6μM; n=3–14; mean±s.e.m. d, Amlodipine (yellow sticks) bound to CaVAb. S5 and S6 helices in ribbons; residues surrounding amlodipine in sticks. e, Nimodipine bound to CaVAb. f, UK-59811 bound to CaVAb. Anomalous scattering density (3σ, green mesh) for Br in UK-59811. g, DMPC lipid in the drug-free dihydropyridine-binding site in yellow sticks. d e g Y195 I199 F167 Y168 F171 TM6 TM6 TM5 Y195 I199 F171 F167 Y168 TM6 TM6 TM5 W195 I199 F171 F167 Y168 F203 M137 F141 TM6 TM6 TM5 a b f Nimodipine UK-59811 DMPC c Amlodipine TM6 TM6 F171 TM5 F167 Y168 Y195 I199 1 μM nimodipine 10 ms 0.5 nA . 100 102 104 Nimodipine (nM) O O O O N H N O O O + – 0.25 0.50 0.75 1.00 0 Norm. current 0.25 0.50 0.75 1.00 0 Norm. current 0 5 10 15 20 Number of pulses Ca I199S VAb © 2016 Macmillan Publishers Limited, part of Springer Nature. All rights reserved