正在加载图片...
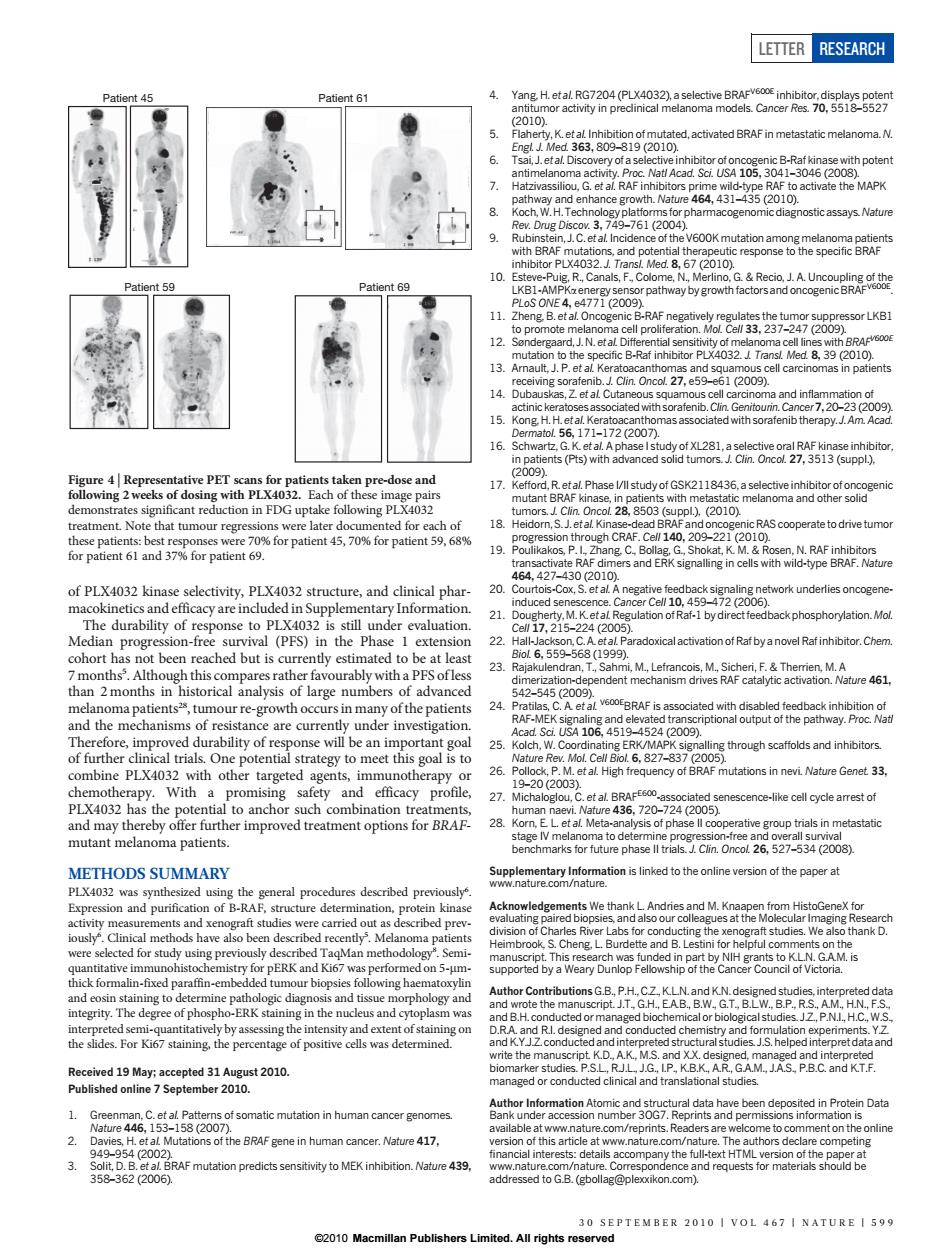
LETTER RESEARCH (P) tatic melanoma N 6. ith poten 8. roh stic assavs Nature 59 t69 oE477209 38,27249288orLk8 13 39 (2010) and s 1200 PET 2118436,a selectiv pl.2010 Note that tu bur regre ns were late 45,70%for patient9. CRAF.C hang d-eAN r Cel 1045372200) derlies oncogen 21 otRat-1 b se to PIX 3 22 lactivation of Rat by a novel Rat inhibitor.Chen cohort hasn t been reached but is cu ently estimated to be at least 3 dran. 5425452089 urre-growtn occu 24 BRAF5为 se will be an im 4106 portant go 25 combine PLX4032 with other targeted agents,imm t al High fre n nevi Nature Genet 33 20(20 PLX4032 an 27 Cet a720-724(2005) cell cycle arrest of 28 ea5ptietnmetasa6 arks tor future phase Cun Oncot 26.5273(2008) METHODS SUMMARY splemetanlhinmaionisnkedtotheoninevesionothepapgra PLX403 zed u dures described p at p ing the xe thank D. ERK a formaln-t pret dataan tion in human cancer get Fgene in human cancer.Natue 417 nd recof PLX4032 kinase selectivity, PLX4032 structure, and clinical pharmacokinetics and efficacy are included in Supplementary Information. The durability of response to PLX4032 is still under evaluation. Median progression-free survival (PFS) in the Phase 1 extension cohort has not been reached but is currently estimated to be at least 7 months5 . Although this compares rather favourably with a PFS of less than 2 months in historical analysis of large numbers of advanced melanoma patients28, tumour re-growth occurs in many of the patients and the mechanisms of resistance are currently under investigation. Therefore, improved durability of response will be an important goal of further clinical trials. One potential strategy to meet this goal is to combine PLX4032 with other targeted agents, immunotherapy or chemotherapy. With a promising safety and efficacy profile, PLX4032 has the potential to anchor such combination treatments, and may thereby offer further improved treatment options for BRAFmutant melanoma patients. METHODS SUMMARY PLX4032 was synthesized using the general procedures described previously6 . Expression and purification of B-RAF, structure determination, protein kinase activity measurements and xenograft studies were carried out as described previously6 . Clinical methods have also been described recently5 . Melanoma patients were selected for study using previously described TaqMan methodology8 . Semiquantitative immunohistochemistry for pERK and Ki67 was performed on 5-mmthick formalin-fixed paraffin-embedded tumour biopsies following haematoxylin and eosin staining to determine pathologic diagnosis and tissue morphology and integrity. The degree of phospho-ERK staining in the nucleus and cytoplasm was interpreted semi-quantitatively by assessing the intensity and extent of staining on the slides. For Ki67 staining, the percentage of positive cells was determined. Received 19 May; accepted 31 August 2010. Published online 7 September 2010. 1. Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007). 2. Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). 3. Solit, D. B. et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439, 358–362 (2006). 4. Yang, H. et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 70, 5518–5527 (2010). 5. Flaherty, K. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010). 6. Tsai, J. et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl Acad. Sci. USA 105, 3041–3046 (2008). 7. Hatzivassiliou, G. et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010). 8. Koch, W. H. Technology platforms for pharmacogenomic diagnostic assays. Nature Rev. Drug Discov. 3, 749–761 (2004). 9. Rubinstein, J. C. et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J. Transl. Med. 8, 67 (2010). 10. Esteve-Puig, R., Canals, F., Colome, N., Merlino, G. & Recio, J. A. Uncoupling of the LKB1-AMPKa energy sensor pathway by growth factors and oncogenic BRAFV600E. PLoS ONE 4, e4771 (2009). 11. Zheng, B. et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell 33, 237–247 (2009). 12. Søndergaard, J. N. et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific B-Raf inhibitor PLX4032. J. Transl. Med. 8, 39 (2010). 13. Arnault, J. P. et al. Keratoacanthomas and squamous cell carcinomas in patients receiving sorafenib. J. Clin. Oncol. 27, e59–e61 (2009). 14. Dubauskas, Z. et al. Cutaneous squamous cell carcinoma and inflammation of actinic keratoses associated with sorafenib.Clin. Genitourin.Cancer7,20–23 (2009). 15. Kong, H. H. et al. Keratoacanthomas associated with sorafenib therapy. J. Am. Acad. Dermatol. 56, 171–172 (2007). 16. Schwartz, G. K. et al. A phase I study of XL281, a selective oral RAF kinase inhibitor, in patients (Pts) with advanced solid tumors. J. Clin. Oncol. 27, 3513 (suppl.), (2009). 17. Kefford, R. et al.Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J. Clin. Oncol. 28, 8503 (suppl.), (2010). 18. Heidorn, S. J. et al.Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140, 209–221 (2010). 19. Poulikakos, P. I., Zhang, C., Bollag, G., Shokat, K. M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010). 20. Courtois-Cox, S. et al. A negative feedback signaling network underlies oncogeneinduced senescence. Cancer Cell 10, 459–472 (2006). 21. Dougherty, M. K. et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17, 215–224 (2005). 22. Hall-Jackson, C. A. et al. Paradoxical activation of Raf by a novel Raf inhibitor.Chem. Biol. 6, 559–568 (1999). 23. Rajakulendran, T., Sahmi, M., Lefrancois, M., Sicheri, F. & Therrien, M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). 24. Pratilas, C. A. et al. V600EBRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl Acad. Sci. USA 106, 4519–4524 (2009). 25. Kolch, W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nature Rev. Mol. Cell Biol. 6, 827–837 (2005). 26. Pollock, P. M. et al. High frequency of BRAF mutations in nevi. Nature Genet. 33, 19–20 (2003). 27. Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005). 28. Korn, E. L. et al. Meta-analysis of phase II cooperative group trials in metastatic stage IV melanoma to determine progression-free and overall survival benchmarks for future phase II trials. J. Clin. Oncol. 26, 527–534 (2008). Supplementary Information is linked to the online version of the paper at www.nature.com/nature. Acknowledgements We thank L. Andries and M. Knaapen from HistoGeneX for evaluating paired biopsies, and also our colleagues at the Molecular Imaging Research division of Charles River Labs for conducting the xenograft studies. We also thank D. Heimbrook, S. Cheng, L. Burdette and B. Lestini for helpful comments on the manuscript. This research was funded in part by NIH grants to K.L.N. G.A.M. is supported by a Weary Dunlop Fellowship of the Cancer Council of Victoria. Author Contributions G.B., P.H., C.Z., K.L.N. and K.N. designed studies, interpreted data and wrote the manuscript. J.T., G.H., E.A.B., B.W., G.T., B.L.W., B.P., R.S., A.M., H.N., F.S., and B.H. conducted or managed biochemical or biological studies. J.Z., P.N.I., H.C., W.S., D.R.A. and R.I. designed and conducted chemistry and formulation experiments. Y.Z. and K.Y.J.Z. conducted and interpreted structural studies. J.S. helped interpret data and write the manuscript. K.D., A.K., M.S. and X.X. designed, managed and interpreted biomarker studies. P.S.L., R.J.L., J.G., I.P., K.B.K., A.R., G.A.M., J.A.S., P.B.C. and K.T.F. managed or conducted clinical and translational studies. Author Information Atomic and structural data have been deposited in Protein Data Bank under accession number 3OG7. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Correspondence and requests for materials should be addressed to G.B. (gbollag@plexxikon.com). Patient 45 Patient 61 Patient 59 Patient 69 Figure 4 | Representative PET scans for patients taken pre-dose and following 2 weeks of dosing with PLX4032. Each of these image pairs demonstrates significant reduction in FDG uptake following PLX4032 treatment. Note that tumour regressions were later documented for each of these patients: best responses were 70% for patient 45, 70% for patient 59, 68% for patient 61 and 37% for patient 69. LETTER RESEARCH 30 SEPTEMBER 2010 | VOL 467 | NATURE | 599 ©2010 Macmillan Publishers Limited. All rights reserved