正在加载图片...
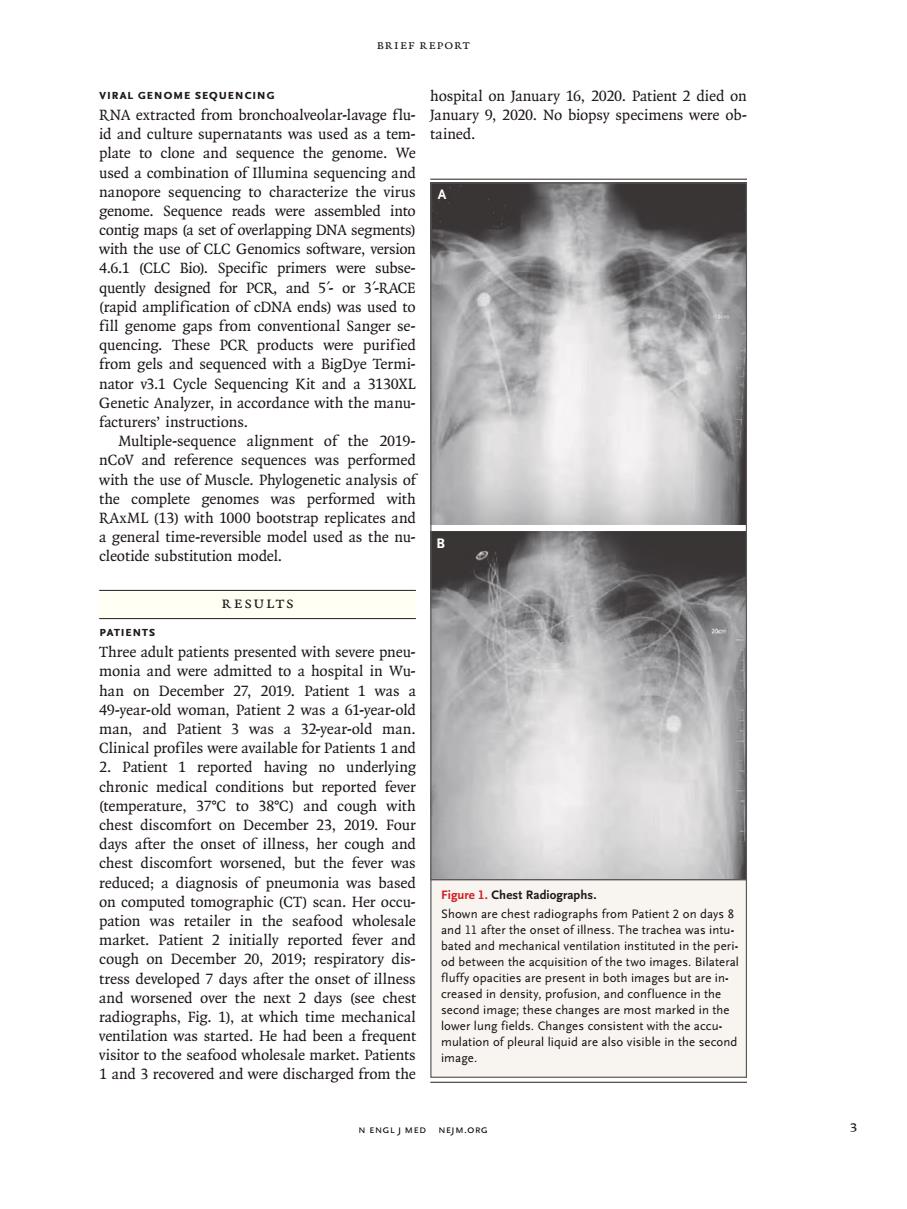
BRIEF REPORT VIRAL GENOME SEQUENCING hospital on lanuary 16.2020.Patient 2 died or RNA extracted from bronchoalveolar-lavage flu-January 9,2020.No biopsy specimens were ob- id and culture supernatants was used as a tem-tained. plate to clone and sequence the genome. used a combination of Illumina sequencing and nanopore sequencing t ing DNA segm with the use of CLC Genomics software.version 4.6.1 (CLC Bio).Specific primers were subse quently designed for PCR, and 5-or 3'-RAC PCR s and sear nator v3.1 Cycle Sequencing Kit and a 3130XI Genetic Analyzer,in accordance with the manu facturers'instructions 2019 eswas performe RAxML (13)wi1OO0bo a general time-reversible model used as the nu cleotide substitution model. RESULTS Three adult ted with s monia and were admitted ro a hospital in wu han on December 27,2019.Patient 1 was a 49-year-old woman,Patient 2 was a 61-year-old 1 an y 38C) and cough chest discomfort on December 23.2019.Fou days after the onset of illness,her cough and chest discomfort worsened,but the fever was reduced;a diagnosis of pneumonia was based Figure 1.Chest Radiographs. ( .Pa d and r r the onse l ventila on i dis tress develoned 7 days after the onset of illnes nt in both es but aren and worsened over the next 2 days (see ches cond i radiographs,Fig.1),at which time mechanica lung was tarte had been a trequen nd w mage. N ENGLJ MED NEJM.ORC n engl j med nejm.org 3 Brief Report Viral Genome Sequencing RNA extracted from bronchoalveolar-lavage fluid and culture supernatants was used as a template to clone and sequence the genome. We used a combination of Illumina sequencing and nanopore sequencing to characterize the virus genome. Sequence reads were assembled into contig maps (a set of overlapping DNA segments) with the use of CLC Genomics software, version 4.6.1 (CLC Bio). Specific primers were subsequently designed for PCR, and 5′- or 3′-RACE (rapid amplification of cDNA ends) was used to fill genome gaps from conventional Sanger sequencing. These PCR products were purified from gels and sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit and a 3130XL Genetic Analyzer, in accordance with the manufacturers’ instructions. Multiple-sequence alignment of the 2019- nCoV and reference sequences was performed with the use of Muscle. Phylogenetic analysis of the complete genomes was performed with RAxML (13) with 1000 bootstrap replicates and a general time-reversible model used as the nucleotide substitution model. Results Patients Three adult patients presented with severe pneumonia and were admitted to a hospital in Wuhan on December 27, 2019. Patient 1 was a 49-year-old woman, Patient 2 was a 61-year-old man, and Patient 3 was a 32-year-old man. Clinical profiles were available for Patients 1 and 2. Patient 1 reported having no underlying chronic medical conditions but reported fever (temperature, 37°C to 38°C) and cough with chest discomfort on December 23, 2019. Four days after the onset of illness, her cough and chest discomfort worsened, but the fever was reduced; a diagnosis of pneumonia was based on computed tomographic (CT) scan. Her occupation was retailer in the seafood wholesale market. Patient 2 initially reported fever and cough on December 20, 2019; respiratory distress developed 7 days after the onset of illness and worsened over the next 2 days (see chest radiographs, Fig. 1), at which time mechanical ventilation was started. He had been a frequent visitor to the seafood wholesale market. Patients 1 and 3 recovered and were discharged from the hospital on January 16, 2020. Patient 2 died on January 9, 2020. No biopsy specimens were obtained. Figure 1. Chest Radiographs. Shown are chest radiographs from Patient 2 on days 8 and 11 after the onset of illness. The trachea was intubated and mechanical ventilation instituted in the period between the acquisition of the two images. Bilateral fluffy opacities are present in both images but are increased in density, profusion, and confluence in the second image; these changes are most marked in the lower lung fields. Changes consistent with the accumulation of pleural liquid are also visible in the second image. A B