正在加载图片...
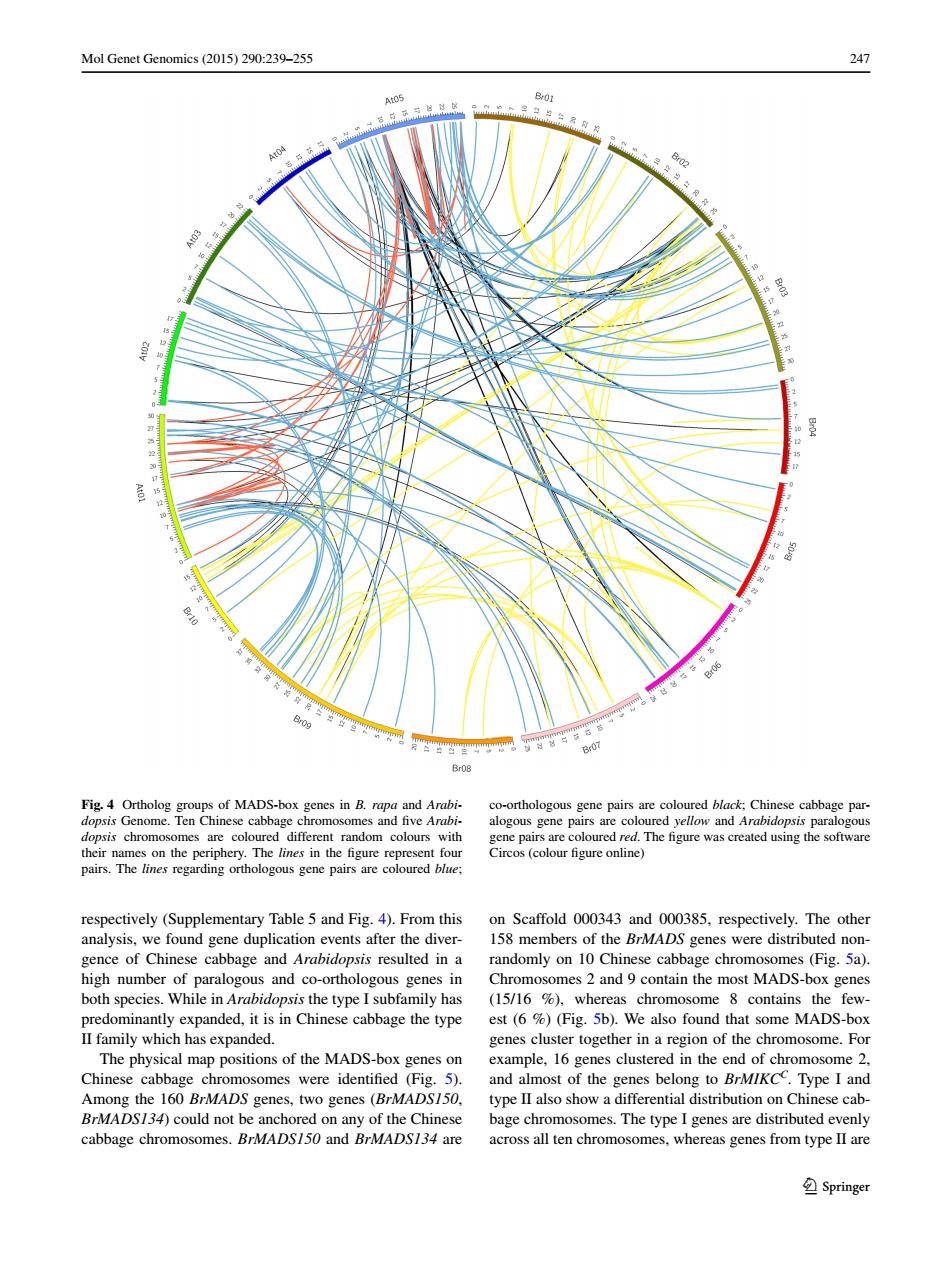
Mol Genet Genomics (2015)290:239-255 247 A0 B01 At04 8e2 罩 81 Br09 Br07 Br08 Fig.4 Ortholog groups of MADS-box genes in B.rapa and Arabi- co-orthologous gene pairs are coloured black:Chinese cabbage par- dopsis Genome.Ten Chinese cabbage chromosomes and five Arabi- alogous gene pairs are coloured yellow and Arabidopsis paralogous dopsis chromosomes are coloured different random colours with gene pairs are coloured red.The figure was created using the software their names on the periphery.The lines in the figure represent four Circos(colour figure online) pairs.The lines regarding orthologous gene pairs are coloured blue; respectively (Supplementary Table 5 and Fig.4).From this on Scaffold 000343 and 000385,respectively.The other analysis,we found gene duplication events after the diver- 158 members of the BrMADS genes were distributed non- gence of Chinese cabbage and Arabidopsis resulted in a randomly on 10 Chinese cabbage chromosomes (Fig.5a). high number of paralogous and co-orthologous genes in Chromosomes 2 and 9 contain the most MADS-box genes both species.While in Arabidopsis the type I subfamily has (15/16 %)whereas chromosome 8 contains the few- predominantly expanded,it is in Chinese cabbage the type est (6 %)(Fig.5b).We also found that some MADS-box II family which has expanded. genes cluster together in a region of the chromosome.For The physical map positions of the MADS-box genes on example,16 genes clustered in the end of chromosome 2. Chinese cabbage chromosomes were identified (Fig.5). and almost of the genes belong to BrMIKCC.Type I and Among the 160 BrMADS genes,two genes (BrMADS150, type II also show a differential distribution on Chinese cab- BrMADS/34)could not be anchored on any of the Chinese bage chromosomes.The type I genes are distributed evenly cabbage chromosomes.BrMADS150 and BrMADS134 are across all ten chromosomes,whereas genes from type II are SpringerMol Genet Genomics (2015) 290:239–255 247 1 3 respectively (Supplementary Table 5 and Fig. 4). From this analysis, we found gene duplication events after the divergence of Chinese cabbage and Arabidopsis resulted in a high number of paralogous and co-orthologous genes in both species. While in Arabidopsis the type I subfamily has predominantly expanded, it is in Chinese cabbage the type II family which has expanded. The physical map positions of the MADS-box genes on Chinese cabbage chromosomes were identified (Fig. 5). Among the 160 BrMADS genes, two genes (BrMADS150, BrMADS134) could not be anchored on any of the Chinese cabbage chromosomes. BrMADS150 and BrMADS134 are on Scaffold 000343 and 000385, respectively. The other 158 members of the BrMADS genes were distributed nonrandomly on 10 Chinese cabbage chromosomes (Fig. 5a). Chromosomes 2 and 9 contain the most MADS-box genes (15/16 %), whereas chromosome 8 contains the fewest (6 %) (Fig. 5b). We also found that some MADS-box genes cluster together in a region of the chromosome. For example, 16 genes clustered in the end of chromosome 2, and almost of the genes belong to BrMIKCC. Type I and type II also show a differential distribution on Chinese cabbage chromosomes. The type I genes are distributed evenly across all ten chromosomes, whereas genes from type II are Fig. 4 Ortholog groups of MADS-box genes in B. rapa and Arabidopsis Genome. Ten Chinese cabbage chromosomes and five Arabidopsis chromosomes are coloured different random colours with their names on the periphery. The lines in the figure represent four pairs. The lines regarding orthologous gene pairs are coloured blue; co-orthologous gene pairs are coloured black; Chinese cabbage paralogous gene pairs are coloured yellow and Arabidopsis paralogous gene pairs are coloured red. The figure was created using the software Circos (colour figure online)