正在加载图片...
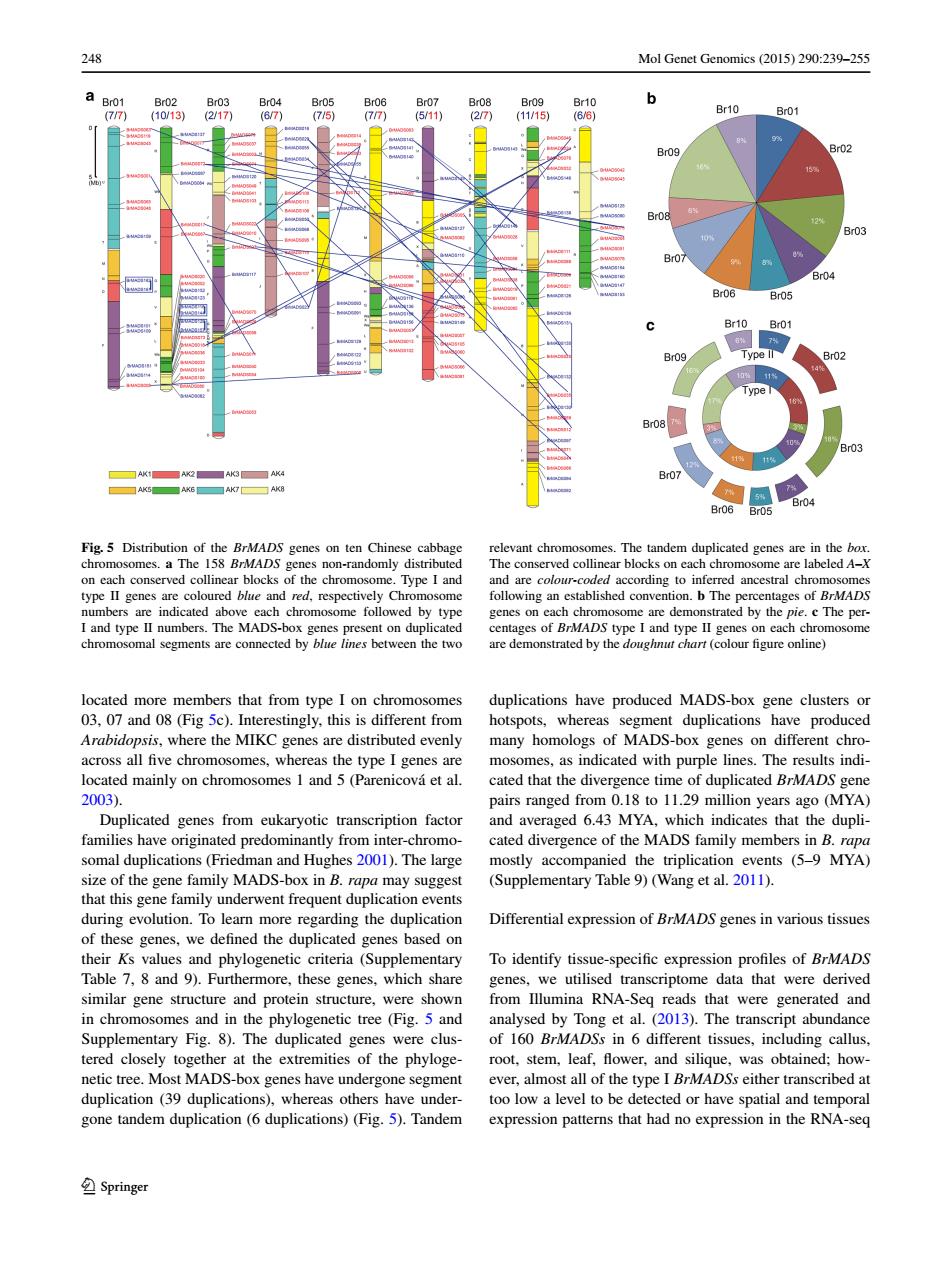
248 Mol Genet Genomics(2015)290:239-255 Br05 Br06 B07 B08 Br10 Br10 77 27 (11/15) (6/6) Br01 Br09 Br02 1 Br03 B07 Br04 Br06 Br05 Br10 Br01 7 Br09 Type ll Br02 Br08 ■AK3☐K Br07 Br04 Br06 Br05 Fig.5 Distribution of the BrMADS genes on ten Chinese cabbage relevant chromosomes.The tandem duplicated genes are in the box. chromosomes.a The 158 BrMADS genes non-randomly distributed The conserved collinear blocks on each chromosome are labeled A-X on each conserved collinear blocks of the chromosome.Type I and and are colour-coded according to inferred ancestral chromosomes type II genes are coloured blue and red,respectively Chromosome following an established convention.b The percentages of BrMADS numbers are indicated above each chromosome followed by type genes on each chromosome are demonstrated by the pie.e The per- I and type II numbers.The MADS-box genes present on duplicated centages of BrMADS type I and type II genes on each chromosome chromosomal segments are connected by blue lines between the two are demonstrated by the doughnut chart (colour figure online) located more members that from type I on chromosomes duplications have produced MADS-box gene clusters or 03,07 and 08(Fig 5c).Interestingly,this is different from hotspots,whereas segment duplications have produced Arabidopsis,where the MIKC genes are distributed evenly many homologs of MADS-box genes on different chro- across all five chromosomes,whereas the type I genes are mosomes,as indicated with purple lines.The results indi- located mainly on chromosomes 1 and 5(Parenicova et al. cated that the divergence time of duplicated BrMADS gene 2003). pairs ranged from 0.18 to 11.29 million years ago (MYA) Duplicated genes from eukaryotic transcription factor and averaged 6.43 MYA,which indicates that the dupli- families have originated predominantly from inter-chromo- cated divergence of the MADS family members in B.rapa somal duplications(Friedman and Hughes 2001).The large mostly accompanied the triplication events (5-9 MYA) size of the gene family MADS-box in B.rapa may suggest (Supplementary Table 9)(Wang et al.2011). that this gene family underwent frequent duplication events during evolution.To learn more regarding the duplication Differential expression of BrMADS genes in various tissues of these genes,we defined the duplicated genes based on their Ks values and phylogenetic criteria (Supplementary To identify tissue-specific expression profiles of BrMADS Table 7,8 and 9).Furthermore,these genes,which share genes,we utilised transcriptome data that were derived similar gene structure and protein structure,were shown from Illumina RNA-Seg reads that were generated and in chromosomes and in the phylogenetic tree (Fig.5 and analysed by Tong et al.(2013).The transcript abundance Supplementary Fig.8).The duplicated genes were clus-of 160 BrMADSs in 6 different tissues,including callus. tered closely together at the extremities of the phyloge-root,stem,leaf,flower,and silique,was obtained;how- netic tree.Most MADS-box genes have undergone segment ever,almost all of the type I BrMADSs either transcribed at duplication (39 duplications),whereas others have under- too low a level to be detected or have spatial and temporal gone tandem duplication (6 duplications)(Fig.5).Tandem expression patterns that had no expression in the RNA-seq Springer248 Mol Genet Genomics (2015) 290:239–255 1 3 located more members that from type I on chromosomes 03, 07 and 08 (Fig 5c). Interestingly, this is different from Arabidopsis, where the MIKC genes are distributed evenly across all five chromosomes, whereas the type I genes are located mainly on chromosomes 1 and 5 (Parenicová et al. 2003). Duplicated genes from eukaryotic transcription factor families have originated predominantly from inter-chromosomal duplications (Friedman and Hughes 2001). The large size of the gene family MADS-box in B. rapa may suggest that this gene family underwent frequent duplication events during evolution. To learn more regarding the duplication of these genes, we defined the duplicated genes based on their Ks values and phylogenetic criteria (Supplementary Table 7, 8 and 9). Furthermore, these genes, which share similar gene structure and protein structure, were shown in chromosomes and in the phylogenetic tree (Fig. 5 and Supplementary Fig. 8). The duplicated genes were clustered closely together at the extremities of the phylogenetic tree. Most MADS-box genes have undergone segment duplication (39 duplications), whereas others have undergone tandem duplication (6 duplications) (Fig. 5). Tandem duplications have produced MADS-box gene clusters or hotspots, whereas segment duplications have produced many homologs of MADS-box genes on different chromosomes, as indicated with purple lines. The results indicated that the divergence time of duplicated BrMADS gene pairs ranged from 0.18 to 11.29 million years ago (MYA) and averaged 6.43 MYA, which indicates that the duplicated divergence of the MADS family members in B. rapa mostly accompanied the triplication events (5–9 MYA) (Supplementary Table 9) (Wang et al. 2011). Differential expression of BrMADS genes in various tissues To identify tissue-specific expression profiles of BrMADS genes, we utilised transcriptome data that were derived from Illumina RNA-Seq reads that were generated and analysed by Tong et al. (2013). The transcript abundance of 160 BrMADSs in 6 different tissues, including callus, root, stem, leaf, flower, and silique, was obtained; however, almost all of the type I BrMADSs either transcribed at too low a level to be detected or have spatial and temporal expression patterns that had no expression in the RNA-seq Fig. 5 Distribution of the BrMADS genes on ten Chinese cabbage chromosomes. a The 158 BrMADS genes non-randomly distributed on each conserved collinear blocks of the chromosome. Type I and type II genes are coloured blue and red, respectively Chromosome numbers are indicated above each chromosome followed by type I and type II numbers. The MADS-box genes present on duplicated chromosomal segments are connected by blue lines between the two relevant chromosomes. The tandem duplicated genes are in the box. The conserved collinear blocks on each chromosome are labeled A–X and are colour-coded according to inferred ancestral chromosomes following an established convention. b The percentages of BrMADS genes on each chromosome are demonstrated by the pie. c The percentages of BrMADS type I and type II genes on each chromosome are demonstrated by the doughnut chart (colour figure online)