正在加载图片...
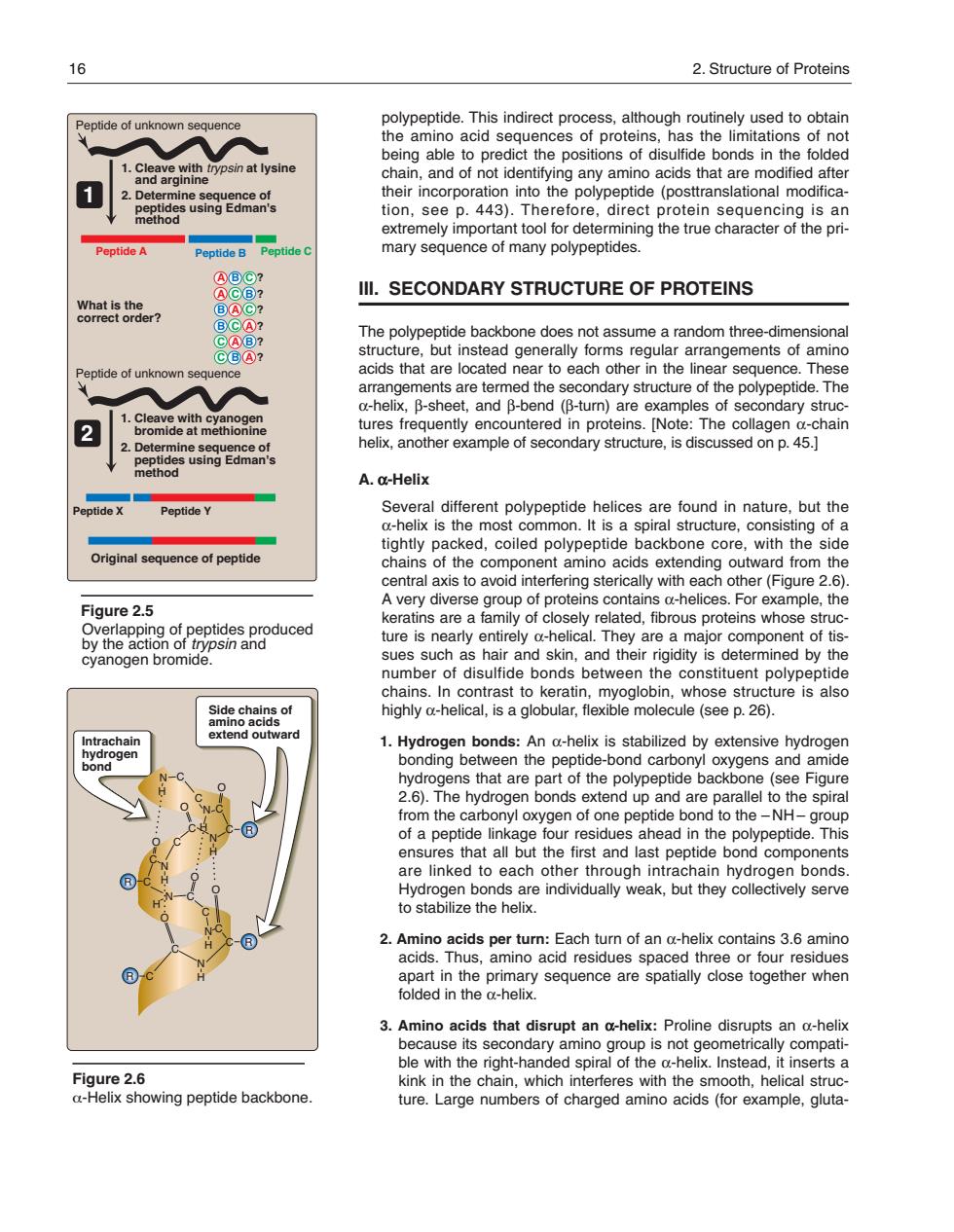
16 2.Structure of Proteins polypeptide.This indirect process,although routinely used to obtain tide of unknown the amino acid sequences of proteins,has the limitations of not ict the positions of dis ds in the fo osin at lysine 1 eptide(posttranslational modifica tion,see p.443).Therefore,direct protein sequencing is an extremely important tool for determining the true character of the pri Peptide A Peptide B Peptide mary sequence of many polypeptides Ill.SECONDARY STRUCTURE OF PROTEINS what is the? me a ra n thre aocuoag0etaad0earo2aeohenenero6nec2n3 anangerBeshe are termed ne secondary stuciure o the pbolypepude.mne eet,and p -turn are e 2 Thor se A.a-Helix ptide Peptide Y Several different polypeptide helices are found in nature.but the a-helix is the most common.It is a spiral structure,consisting of a Original sequenc of peptide tightly packed,coiled polypeptide ackbone core,with the side chains aoonent mino ac ending o er (F A very diverse group of proteins contains a-helices.For ample.the Figure 2.5 keratins are a family of closely related,fibrous proteins whose struc ure is rely d- are major cyanogen bromide omponen chains.In contrast to keratin,myoglobin,whose structure is also highly a-helical,is a globular,flexible molecule(see p.26). An a-helix is stabilized by extensivehy ns that are art ot the eptide back (see Fiou 2.6.The hydrogen bonds extend up and are parallel to the spiral rom the carbonyl oxygen of one pep tide ond to the -NH group but the first and las are linked to each other through intrachain hydrogen bonds. gse2athorendrduayweak,btheyolecneysone apart in the primary sequence are spatially close together when folded in the a-helix 3.Amino acids that disrupt an a-helix:Proline disrupts an a-heli le its dary amino oup is no Figure 2.6 kinkn the chain which interferes with the smooth.helical struc a-Helix showing peptide backbone. ture.Large numbers of charged amino acids(for example,gluta- polypeptide. This indirect process, although routinely used to obtain the amino acid sequences of proteins, has the limitations of not being able to predict the positions of disulfide bonds in the folded chain, and of not identifying any amino acids that are modified after their incorporation into the polypeptide (posttranslational modification, see p. 443). Therefore, direct protein sequencing is an extremely important tool for determining the true character of the primary sequence of many polypeptides. III. SECONDARY STRUCTURE OF PROTEINS The polypeptide backbone does not assume a random three-dimensional structure, but instead generally forms regular arrangements of amino acids that are located near to each other in the linear sequence. These arrangements are termed the secondary structure of the polypeptide. The α-helix, β-sheet, and β-bend (β-turn) are examples of secondary structures frequently encountered in proteins. [Note: The collagen α-chain helix, another example of secondary structure, is discussed on p. 45.] A. α-Helix Several different polypeptide helices are found in nature, but the α-helix is the most common. It is a spiral structure, consisting of a tightly packed, coiled polypeptide backbone core, with the side chains of the component amino acids extending outward from the central axis to avoid interfering sterically with each other (Figure 2.6). A very diverse group of proteins contains α-helices. For example, the keratins are a family of closely related, fibrous proteins whose structure is nearly entirely α-helical. They are a major component of tissues such as hair and skin, and their rigidity is determined by the number of disulfide bonds between the constituent polypeptide chains. In contrast to keratin, myoglobin, whose structure is also highly α-helical, is a globular, flexible molecule (see p. 26). 1. Hydrogen bonds: An α-helix is stabilized by extensive hydrogen bonding between the peptide-bond carbonyl oxygens and amide hydrogens that are part of the polypeptide backbone (see Figure 2.6). The hydrogen bonds extend up and are parallel to the spiral from the carbonyl oxygen of one peptide bond to the – NH – group of a peptide linkage four residues ahead in the polypeptide. This ensures that all but the first and last peptide bond components are linked to each other through intrachain hydrogen bonds. Hydrogen bonds are individually weak, but they collectively serve to stabilize the helix. 2. Amino acids per turn: Each turn of an α-helix contains 3.6 amino acids. Thus, amino acid residues spaced three or four residues apart in the primary sequence are spatially close together when folded in the α-helix. 3. Amino acids that disrupt an α-helix: Proline disrupts an α-helix because its secondary amino group is not geometrically compatible with the right-handed spiral of the α-helix. Instead, it inserts a kink in the chain, which interferes with the smooth, helical structure. Large numbers of charged amino acids (for example, glutaFigure 2.6 α-Helix showing peptide backbone. Side chains of amino acids extend outward Intrachain hydrogen bond N H C O C O N C C N H H O C C C N H O C C O O H N C C N H N H R C C C R R R 16 2. Structure of Proteins Figure 2.5 1. Cleave with trypsin at lysine and arginine Peptide of unknown sequence 2. Determine sequence of peptides using Edman's method What is the correct order? Peptide A Peptide B Peptide X Peptide Y Peptide C 1. Cleave with cyanogen bromide at methionine 2. Determine sequence of peptides using Edman's method 1 2 Original sequence of peptide A B C ? A C B ? B A C ? B C A ? C A B ? C B A ? Overlapping of peptides produced by the action of trypsin and cyanogen bromide. Peptide of unknown sequence 168397_P013-024.qxd7.0:02 Protein structure 5-20-04 2010.4.4 11:31 AM Page 16