正在加载图片...
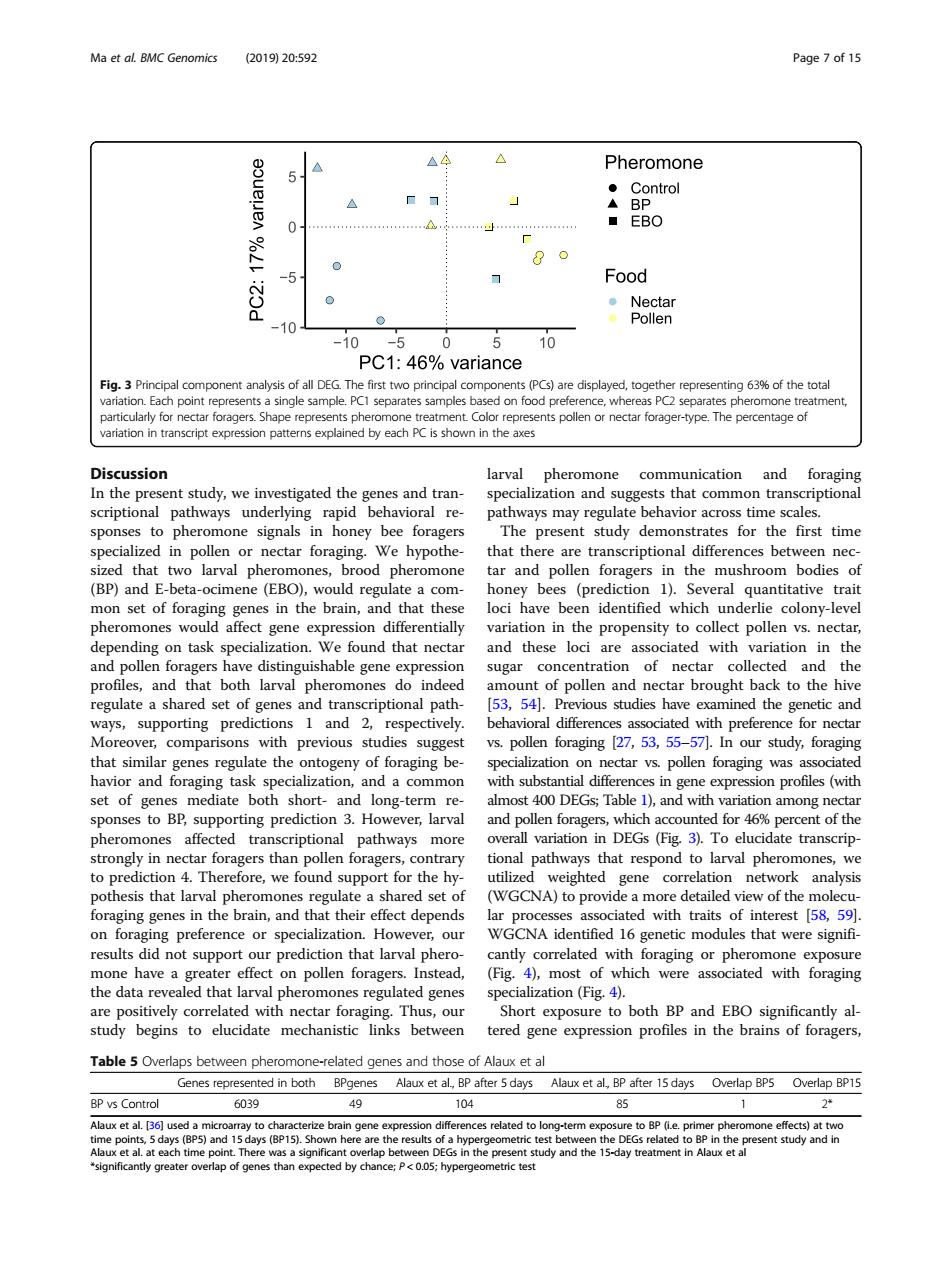
Ma et aL BMC Genomics 2019)20:592 Page7 of 15 △A Pheromone 0 Food 0 10 PC1:46%variance or nectar forager-type.The percentageo Discussion pheromone communication and n the present study,weinv alization and s tim that d that (BP)and larval pheromones,brood pheromone and pollen foragers in the mushroom bodies of ocimene (EBO),woul a regu com era tha quantitative ion diff ntially ariation in the】 epending on task specialization.We found that nectar variation in the sion sugar concentration of nectar collected and the and larval ph en an brought back to the hive for necta Moreover,comparisons with previous studies suggest vs.pollen foraging 27,53,55-57.In our study,foraging that similar genes regulate the ontogeny of foraging be pecialization on nectar vs pollen foraging was a or and in gen xpre (wi and pollen foragers,which ounted for 46% of the pheromones affected transcriptic onal pathways more overall variation in DEGs (Fig 3).To elucidate transcrip trongly in nectar foragers than poll orage onal pathw respond t larval pher mone to pre that lar we (WGCNA)to ene nds ciated with traits of interest [58,59] specialization.Howeve ntified 16 genetic modules that were signifi esult that larva phero hich ith osur raging ialization (Fi Short exposure to both BP and EBO significantly al study begins to elucidate between tered gen expression protiles in the brains of foragers Table5 Overlaps be wee pheromon related genes and those of Alaux et al enes repre d in both after 5 days Alaux et al.B et al.136l used a micr ray to characterze br -pp d ment in Ala Discussion In the present study, we investigated the genes and transcriptional pathways underlying rapid behavioral responses to pheromone signals in honey bee foragers specialized in pollen or nectar foraging. We hypothesized that two larval pheromones, brood pheromone (BP) and E-beta-ocimene (EBO), would regulate a common set of foraging genes in the brain, and that these pheromones would affect gene expression differentially depending on task specialization. We found that nectar and pollen foragers have distinguishable gene expression profiles, and that both larval pheromones do indeed regulate a shared set of genes and transcriptional pathways, supporting predictions 1 and 2, respectively. Moreover, comparisons with previous studies suggest that similar genes regulate the ontogeny of foraging behavior and foraging task specialization, and a common set of genes mediate both short- and long-term responses to BP, supporting prediction 3. However, larval pheromones affected transcriptional pathways more strongly in nectar foragers than pollen foragers, contrary to prediction 4. Therefore, we found support for the hypothesis that larval pheromones regulate a shared set of foraging genes in the brain, and that their effect depends on foraging preference or specialization. However, our results did not support our prediction that larval pheromone have a greater effect on pollen foragers. Instead, the data revealed that larval pheromones regulated genes are positively correlated with nectar foraging. Thus, our study begins to elucidate mechanistic links between larval pheromone communication and foraging specialization and suggests that common transcriptional pathways may regulate behavior across time scales. The present study demonstrates for the first time that there are transcriptional differences between nectar and pollen foragers in the mushroom bodies of honey bees (prediction 1). Several quantitative trait loci have been identified which underlie colony-level variation in the propensity to collect pollen vs. nectar, and these loci are associated with variation in the sugar concentration of nectar collected and the amount of pollen and nectar brought back to the hive [53, 54]. Previous studies have examined the genetic and behavioral differences associated with preference for nectar vs. pollen foraging [27, 53, 55–57]. In our study, foraging specialization on nectar vs. pollen foraging was associated with substantial differences in gene expression profiles (with almost 400 DEGs; Table 1), and with variation among nectar and pollen foragers, which accounted for 46% percent of the overall variation in DEGs (Fig. 3). To elucidate transcriptional pathways that respond to larval pheromones, we utilized weighted gene correlation network analysis (WGCNA) to provide a more detailed view of the molecular processes associated with traits of interest [58, 59]. WGCNA identified 16 genetic modules that were significantly correlated with foraging or pheromone exposure (Fig. 4), most of which were associated with foraging specialization (Fig. 4). Short exposure to both BP and EBO significantly altered gene expression profiles in the brains of foragers, Fig. 3 Principal component analysis of all DEG. The first two principal components (PCs) are displayed, together representing 63% of the total variation. Each point represents a single sample. PC1 separates samples based on food preference, whereas PC2 separates pheromone treatment, particularly for nectar foragers. Shape represents pheromone treatment. Color represents pollen or nectar forager-type. The percentage of variation in transcript expression patterns explained by each PC is shown in the axes Table 5 Overlaps between pheromone-related genes and those of Alaux et al Genes represented in both BPgenes Alaux et al., BP after 5 days Alaux et al., BP after 15 days Overlap BP5 Overlap BP15 BP vs Control 6039 49 104 85 1 2* Alaux et al. [36] used a microarray to characterize brain gene expression differences related to long-term exposure to BP (i.e. primer pheromone effects) at two time points, 5 days (BP5) and 15 days (BP15). Shown here are the results of a hypergeometric test between the DEGs related to BP in the present study and in Alaux et al. at each time point. There was a significant overlap between DEGs in the present study and the 15-day treatment in Alaux et al *significantly greater overlap of genes than expected by chance; P < 0.05; hypergeometric test Ma et al. BMC Genomics (2019) 20:592 Page 7 of 15