正在加载图片...
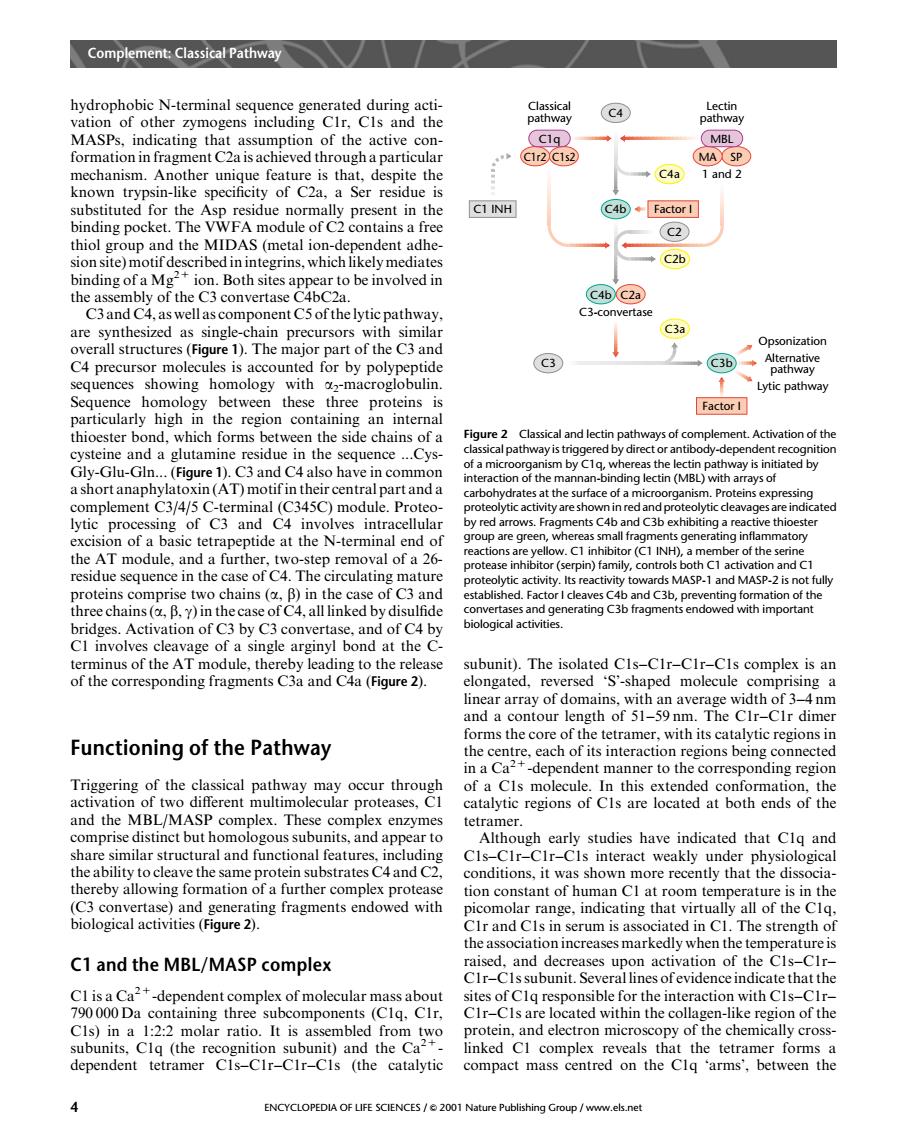
hydrophobic N-terminal sequence generated during acti- vation of other zymogens including CIr.Cls and the of the active con- th →C4a substituted for the Asp residue normally present in the C1INH C4b◆Factor I binding pocket. m0d1 of C contains a (C2) hialgm the (metal which likely media ◆C2b hindingofa mo c46c2 C3an ay C3a 1 Th maior part of the C3 and C4 precursor molecules is accounted for by polype tide sequences showing homology with acroglobulin Lytic pathway betwe thes three proteins Factor I hich fo cide ch Figure 2 Classical and lectin pathways of c mplement Activation of the cysteine and a glutamine residue in the sequence f a m ctin vay is ini (Figure 1).C3 and C4 also have in common SA(AT)m arboh drat at the surface of a m sin ment of C3 and CA c)m red a ents C4b and C xhibiti g a rea ve thi action or (C1 INH) of the sen e in the towards MASP-1 and MASP-2 is no bridges.Activation of C3 by C3 convertase.and of C4 by C1 involves cleavage of a single arginyl bond at the C terminus of the A odule,thereby le ase .The isolat -CIs complex is an ng fragments ed Cis-CIr-CIr ar y of domain with width of um and a contour length of 51-59nm.The CIr-CIr dime Functioning of the Pathway rms the core of the tetramer,with its catalytic regions in the of a Cls molecule In this extended c h ion catalytic regions of Cls are located at both ends of the and the MBL/MASH x enzymes tetramer. us su app studies have indicated that the ability to cleave the same protein substrates C4 and C2. conditions.it was shown more recently that the dissocia thereby allowing formation of a further complex protease tion constant of human Cl at room ter mperature is in the 6a2 ing fragments endowed with picomolar range,indicating that virtu lly all of the Clq serum is as h C1 and the MBL/MASP complex ed and de of the CIr-CIssubunit.Several linesofevidenceindicatethat the -dependent complex of molecular ma ass about containing three sites of Clq respon ated wit for the interaction with CIs-CIr the co th (the rat of the ke region c subunits.Clq nit)and the Co inked Cl complex reveals that the tetra mer forms a dependent tetramer Cis-CIr-CIr-CIs (the catalytic compact mass centred on the Clg 'arms'.between the 4 ENCYCLOPEDIA OF LIFE SCIENCES/2001 Nahydrophobic N-terminal sequence generated during activation of other zymogens including C1r, C1s and the MASPs, indicating that assumption of the active conformation in fragment C2a is achieved through a particular mechanism. Another unique feature is that, despite the known trypsin-like specificity of C2a, a Ser residue is substituted for the Asp residue normally present in the binding pocket. The VWFA module of C2 contains a free thiol group and the MIDAS (metal ion-dependent adhesion site) motif described in integrins, which likely mediates binding of a Mg2+ ion. Both sites appear to be involved in the assembly of the C3 convertase C4bC2a. C3 and C4, as well as component C5 of the lytic pathway, are synthesized as single-chain precursors with similar overall structures (Figure 1). The major part of the C3 and C4 precursor molecules is accounted for by polypeptide sequences showing homology with a2-macroglobulin. Sequence homology between these three proteins is particularly high in the region containing an internal thioester bond, which forms between the side chains of a cysteine and a glutamine residue in the sequence ...CysGly-Glu-Gln... (Figure 1). C3 and C4 also have in common a short anaphylatoxin (AT) motif in their central part and a complement C3/4/5 C-terminal (C345C) module. Proteolytic processing of C3 and C4 involves intracellular excision of a basic tetrapeptide at the N-terminal end of the AT module, and a further, two-step removal of a 26- residue sequence in the case of C4. The circulating mature proteins comprise two chains (a, b) in the case of C3 and three chains (a, b, g) in the case of C4, all linked by disulfide bridges. Activation of C3 by C3 convertase, and of C4 by C1 involves cleavage of a single arginyl bond at the Cterminus of the AT module, thereby leading to the release of the corresponding fragments C3a and C4a (Figure 2). Functioning of the Pathway Triggering of the classical pathway may occur through activation of two different multimolecular proteases, C1 and the MBL/MASP complex. These complex enzymes comprise distinct but homologous subunits, and appear to share similar structural and functional features, including the ability to cleave the same protein substrates C4 and C2, thereby allowing formation of a further complex protease (C3 convertase) and generating fragments endowed with biological activities (Figure 2). C1 and the MBL/MASP complex C1 is a Ca2+-dependent complex of molecular mass about 790 000 Da containing three subcomponents (C1q, C1r, C1s) in a 1:2:2 molar ratio. It is assembled from two subunits, C1q (the recognition subunit) and the Ca2+- dependent tetramer C1s–C1r–C1r–C1s (the catalytic subunit). The isolated C1s–C1r–C1r–C1s complex is an elongated, reversed ‘S’-shaped molecule comprising a linear array of domains, with an average width of 3–4 nm and a contour length of 51–59 nm. The C1r–C1r dimer forms the core of the tetramer, with its catalytic regions in the centre, each of its interaction regions being connected in a Ca2+-dependent manner to the corresponding region of a C1s molecule. In this extended conformation, the catalytic regions of C1s are located at both ends of the tetramer. Although early studies have indicated that C1q and C1s–C1r–C1r–C1s interact weakly under physiological conditions, it was shown more recently that the dissociation constant of human C1 at room temperature is in the picomolar range, indicating that virtually all of the C1q, C1r and C1s in serum is associated in C1. The strength of the association increases markedly when the temperature is raised, and decreases upon activation of the C1s–C1r– C1r–C1s subunit. Several lines of evidence indicate that the sites of C1q responsible for the interaction with C1s–C1r– C1r–C1s are located within the collagen-like region of the protein, and electron microscopy of the chemically crosslinked C1 complex reveals that the tetramer forms a compact mass centred on the C1q ‘arms’, between the Alternative pathway Opsonization Lytic pathway Classical pathway Lectin pathway C4a 1 and 2 C2 C2b C4b C4 C1r2 C1s2 MA SP C1q MBL Factor I C4b C2a C3-convertase C3 C3b Factor I C3a C1 INH Figure 2 Classical and lectin pathways of complement. Activation of the classical pathway is triggered by direct or antibody-dependent recognition of a microorganism by C1q, whereas the lectin pathway is initiated by interaction of the mannan-binding lectin (MBL) with arrays of carbohydrates at the surface of a microorganism. Proteins expressing proteolytic activity are shown in red and proteolytic cleavages are indicated by red arrows. Fragments C4b and C3b exhibiting a reactive thioester group are green, whereas small fragments generating inflammatory reactions are yellow. C1 inhibitor (C1 INH), a member of the serine protease inhibitor (serpin) family, controls both C1 activation and C1 proteolytic activity. Its reactivity towards MASP-1 and MASP-2 is not fully established. Factor I cleaves C4b and C3b, preventing formation of the convertases and generating C3b fragments endowed with important biological activities. Complement: Classical Pathway 4 ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group / www.els.net