
Chapter 11 Phage strategies 清革大当
Chapter 11 Phage strategies

11.1 Introduction 11.2 Lytic development is divided into two periods 11.3 Lytic development is controlled by a cascade 11.4 Functional clustering in phages T7 and T4 11.5 Lambda immediate early and delayed genes are needed for both lysogeny and the lytic cycle 11.6 The lytic cycle depends on antitermination 11.7 Lysogeny is maintained by repressor protein 11.8 Repressor maintains an autogenous circuit 11.9 The repressor and its operators define the immunity region 11.10 The DNA-binding form ofrepressor is a dimer 11.11 Repressor uses a helix-turn-helix motif to bind DNA 11.12 Repressor dimers bind cooperatively to the operator 11.13 Repressor at OR2 interacts with RNA polymerase at PRM 11.14 The cIl and cIll genes are needed to establish lysogeny 11.15 PRE is a poor promoter that requires cll protein 11.16 Lysogeny requires several events 11.17The cro repressor is needed for lytic infection 11.18 What determines the balance between lysogenic and the lytic cycle? 情菜大学
11.1 Introduction 11.2 Lytic development is divided into two periods 11.3 Lytic development is controlled by a cascade 11.4 Functional clustering in phages T7 and T4 11.5 Lambda immediate early and delayed genes are needed for both lysogeny and the lytic cycle 11.6 The lytic cycle depends on antitermination 11.7 Lysogeny is maintained by repressor protein 11.8 Repressor maintains an autogenous circuit 11.9 The repressor and its operators define the immunity region 11.10 The DNA-binding form of repressor is a dimer 11.11 Repressor uses a helix-turn-helix motif to bind DNA 11.12 Repressor dimers bind cooperatively to the operator 11.13 Repressor at OR2 interacts with RNA polymerase at PRM 11.14 The cII and cIII genes are needed to establish lysogeny 11.15 PRE is a poor promoter that requires cII protein 11.16 Lysogeny requires several events 11.17 The cro repressor is needed for lytic infection 11.18 What determines the balance between lysogenic and the lytic cycle?
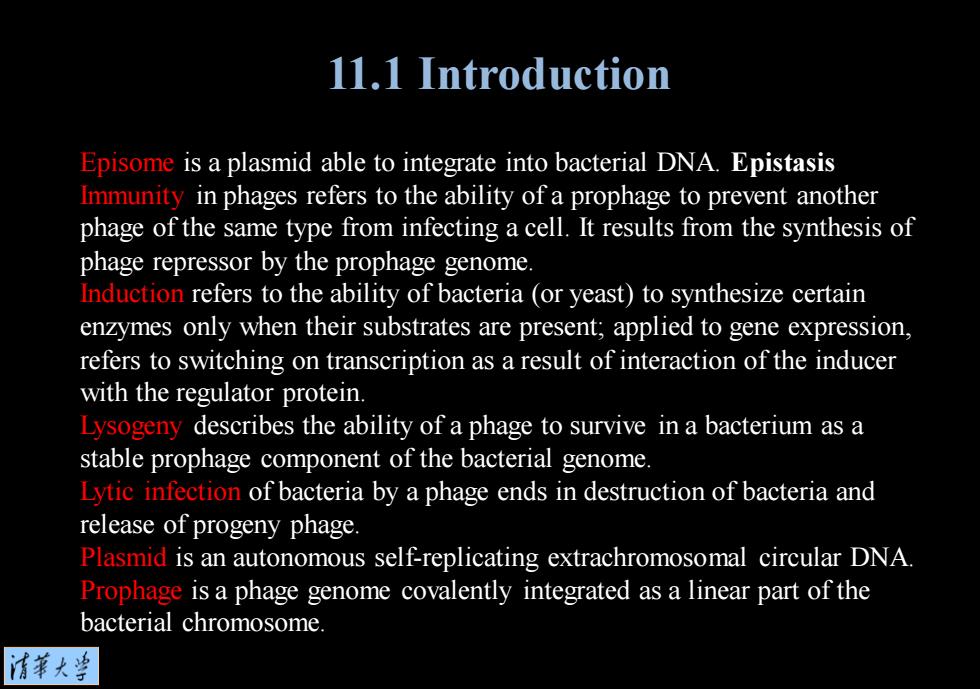
11.1 Introduction Episome is a plasmid able to integrate into bacterial DNA.Epistasis Immunity in phages refers to the ability of a prophage to prevent another phage of the same type from infecting a cell.It results from the synthesis of phage repressor by the prophage genome. Induction refers to the ability of bacteria (or yeast)to synthesize certain enzymes only when their substrates are present;applied to gene expression, refers to switching on transcription as a result of interaction of the inducer with the regulator protein. Lysogeny describes the ability of a phage to survive in a bacterium as a stable prophage component of the bacterial genome. Lytic infection of bacteria by a phage ends in destruction of bacteria and release of progeny phage. Plasmid is an autonomous self-replicating extrachromosomal circular DNA Prophage is a phage genome covalently integrated as a linear part of the bacterial chromosome 清菜大当
Episome is a plasmid able to integrate into bacterial DNA. Epistasis Immunity in phages refers to the ability of a prophage to prevent another phage of the same type from infecting a cell. It results from the synthesis of phage repressor by the prophage genome. Induction refers to the ability of bacteria (or yeast) to synthesize certain enzymes only when their substrates are present; applied to gene expression, refers to switching on transcription as a result of interaction of the inducer with the regulator protein. Lysogeny describes the ability of a phage to survive in a bacterium as a stable prophage component of the bacterial genome. Lytic infection of bacteria by a phage ends in destruction of bacteria and release of progeny phage. Plasmid is an autonomous self-replicating extrachromosomal circular DNA. Prophage is a phage genome covalently integrated as a linear part of the bacterial chromosome. 11.1 Introduction
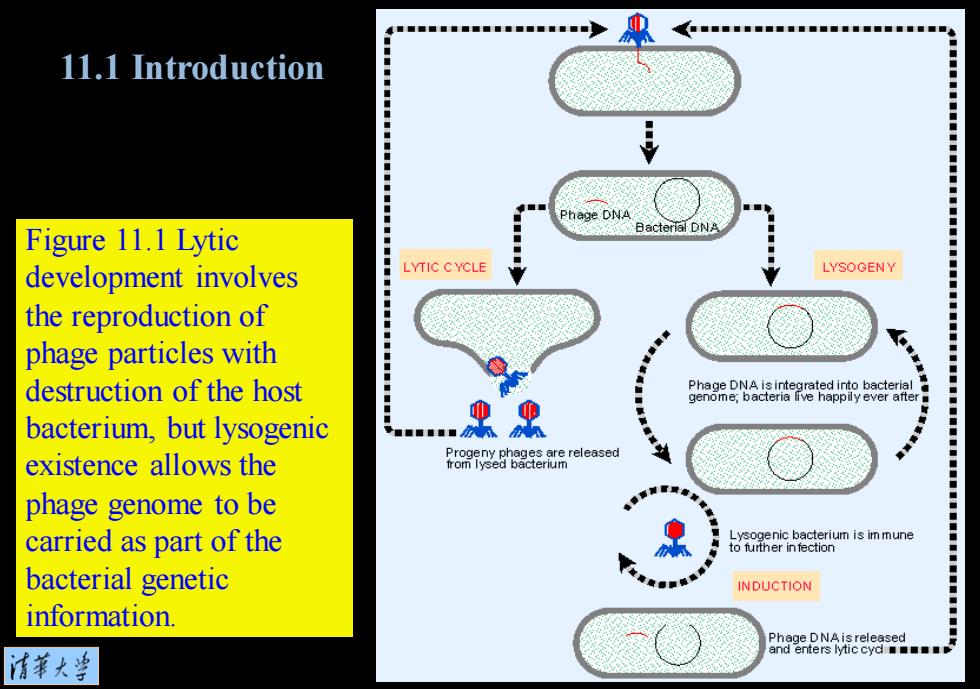
11.1 Introduction Figure 11.1 Lytic development involves LYTIC CYCLE 江” LYSOGEN¥ the reproduction of phage particles with destruction of the host cn ve hpye Phage DNA is integrated into bacterial bacterium,but lysogenic ■■■■■■ existence allows the 6R%838,品released phage genome to be carried as part of the Lysogenic bacterium is im mune to further in fection bacterial genetic INDUCTION information. 清菜大当
Figure 11.1 Lytic development involves the reproduction of phage particles with destruction of the host bacterium, but lysogenic existence allows the phage genome to be carried as part of the bacterial genetic information. 11.1 Introduction
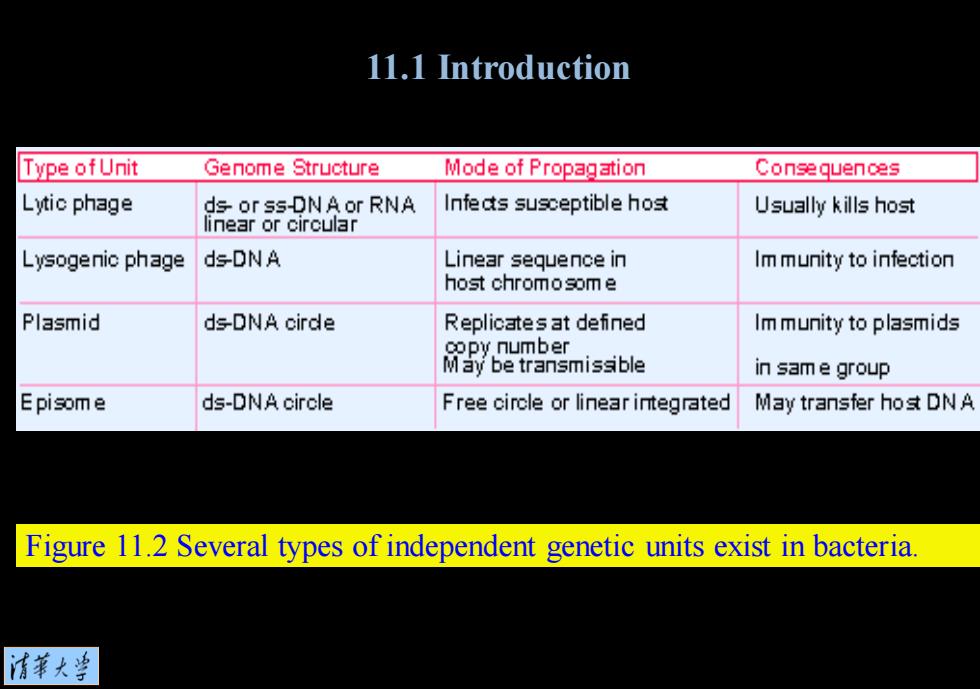
11.1 Introduction Type ofUnit Genome Structure Mode of Propagation Consequences Lytic phage ds or ss-DNA or RNA Infe ats susceptible host Usually kills hos式 linear or circular Lysogenic phage ds-DNA Linear sequence in Im munity to infection host chromosome Plasmid ds-DNA cirde Replicates at defined Im munity to plasmids oopy number May be transmissible in sam e group Episome ds-DNA circle Free circle or linear integrated May transfer ho式DNA Figure 11.2 Several types of independent genetic units exist in bacteria. 清菜大兰
Figure 11.2 Several types of independent genetic units exist in bacteria. 11.1 Introduction

11.2 Lytic development Infection is controlled by a Phage cascade DNA eveiopment ONA Syntnesis are made B酬cation Figure 11.3 Lytic development takes place by Late development producing phage genomes and protein particles that DNA packaged into heads; tails attached are assembled into progeny Lysis phages. erele asoken progery phages 情菜大当
Figure 11.3 Lytic development takes place by producing phage genomes and protein particles that are assembled into progeny phages. 11.2 Lytic development is controlled by a cascade
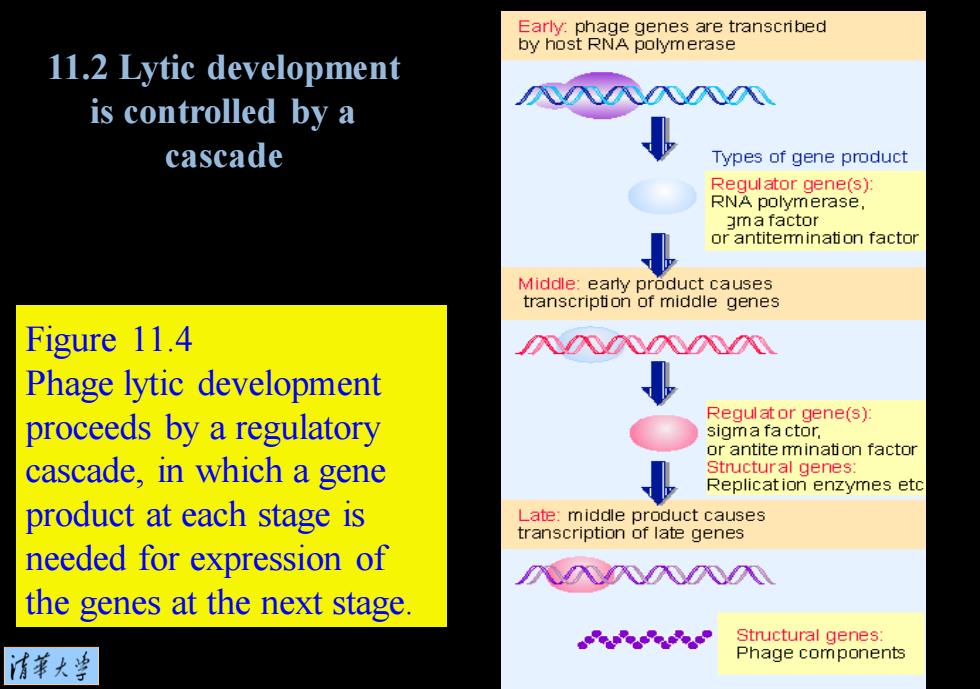
Early:phage genes are transcribed by host RNA polymerase 11.2 Lytic development is controlled by a cascade Types of gene product Requlator gene(s): RNA polymerase, gma factor or antitemmination factor Middle:early product causes transcription of middle genes Figure 11.4 入入入N Phage lytic development Regul at or gene(s): proceeds by a regulatory sigm a fa ctor, or antite mination factor cascade,in which a gene Structural genes Replication enzymes etc product at each stage is Late:middle product causes transcription of late genes needed for expression of 八风入八八 the genes at the next stage. Structural genes: 清第大当 Phage components
Figure 11.4 Phage lytic development proceeds by a regulatory cascade, in which a gene product at each stage is needed for expression of the genes at the next stage. 11.2 Lytic development is controlled by a cascade
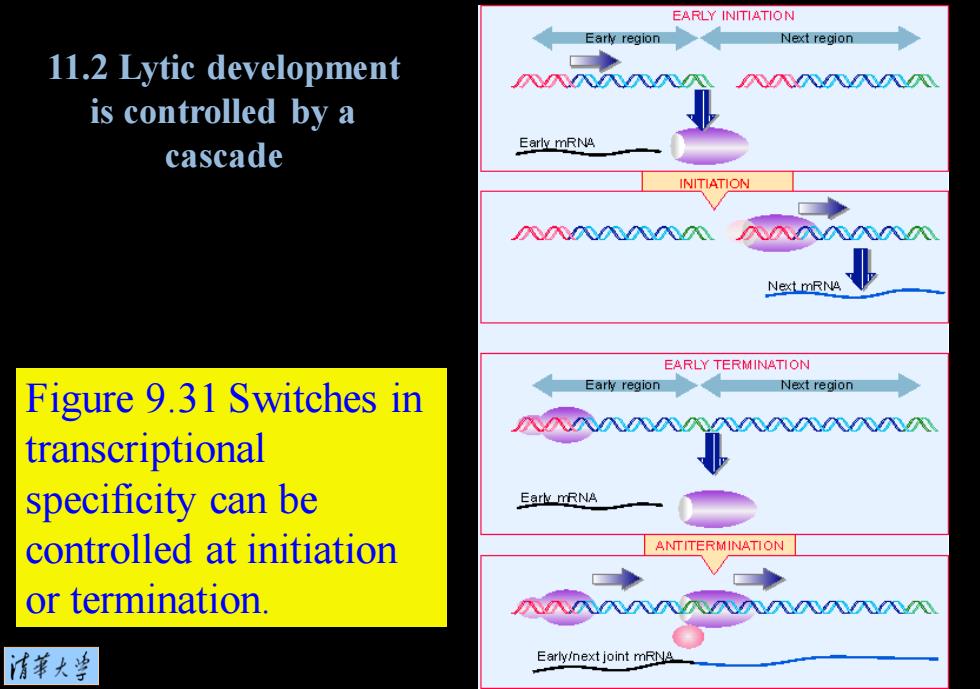
EARLY INITIATION Early region Next region 11.2 Lytic development 入入NNNN is controlled by a Early mRNA cascade INITIATION 八八八0Z02020 八入入入八NN Next mRNA EARLY TERMINATION Figure 9.31 Switches in Early region Next region transcriptional specificity can be Eark mRNA controlled at initiation ANTITERMINATION or termination. 入八NN入八八八入八N 情華大当 Early/next joint mRNA
Figure 9.31 Switches in transcriptional specificity can be controlled at initiation or termination. 11.2 Lytic development is controlled by a cascade

11.3 Functional Class I Class lI Class Ill clustering in phages T7 and T4 5 genes 7 genes 13 genes RNA polymerase ■■■■■■ Host interference Figure 11.5 Phage T7 contains three classes DNA synthesis of genes that are Lysozyme expressed sequentially.The Heads tails genome is ~38 kb DNA maturatior 清第大当
Figure 11.5 Phage T7 contains three classes of genes that are expressed sequentially. The genome is ~38 kb. 11.3 Functional clustering in phages T7 and T4

11.3 Functional clustering in phages T7 and T4 Figure 11.6 late RNA 55 The map ofT4 late RNA 45 tthymidine kinase is circular. de7V endonuclease V 42,43.62,44 io//,ip internal proteins There is DNA polymerase ete. 40 lys ozyme 57 tail fiber extensive head 40 dNMP kinase clustering of 3 sheath terminato 2 head completion genes coding 朗se 50 head completion for 65 head completion 80 5 baseplate plug components of topoisomerase the phage and Okb 6.7.89.10.12 A.日 processes such 160 b aseplate wvedge topoisomerase as DNA ac.13.19.1伦.7 replication,but 3837,363534 head there is also tail fibers 120 dispersion of late RNA 33 格sheath.19tal dTMP synthetase 2 genes coding for a variety of DNA ligase 30 baseplate 54.48 裂0mm0好2斗 enzymatic and 27.51.2626 baseplate plug other functions Essential genes are indicated by numbers.Nonessential genes are identified by letters.Only some representative T4 genes are shown on the map
Essential genes are indicated by numbers. Nonessential genes are identified by letters. Only some representative T4 genes are shown on the map. 11.3 Functional clustering in phages T7 and T4 Figure 11.6 The map of T4 is circular. There is extensive clustering of genes coding for components of the phage and processes such as DNA replication, but there is also dispersion of genes coding for a variety of enzymatic and other functions