CopyrightThe McGraw-Hill Companies,Inc.Permission required for reproduction or display. Chapter 13 Chromatin Structure and Its Effects on Transcription David M.Phillips/Visuals Unlimited
Chapter 13 Chromatin Structure and Its Effects on Transcription
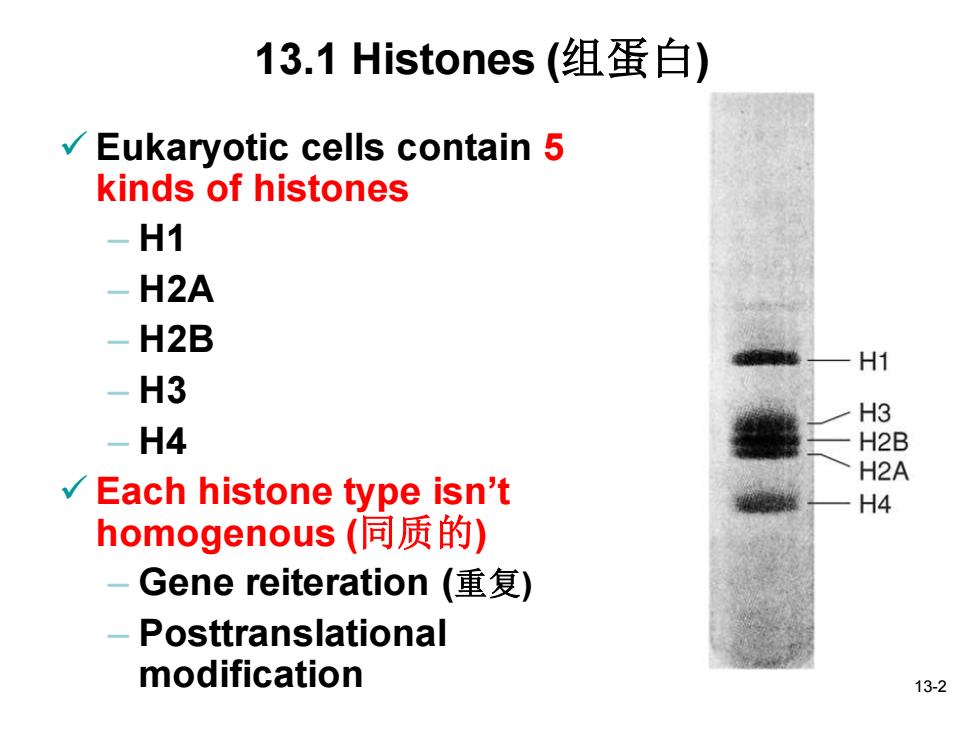
13.1 Histones(组蛋白) / Eukaryotic cells contain 5 kinds of histones -H1 -H2A H2B H1 -H3 H3 -H4 H2B H2A √ Each histone type isn't H4 homogenous(同质的) -Gene reiteration(重复) -Posttranslational modification 13-2
13-2 13.1 Histones (组蛋白) ✓ Eukaryotic cells contain 5 kinds of histones – H1 – H2A – H2B – H3 – H4 ✓ Each histone type isn’t homogenous (同质的) – Gene reiteration (重复) – Posttranslational modification

Properties of Histones Abundant proteins whose mass in nuclei nearly equals that of dNA Pronounced positive charge at neutral pH Most are well-conserved from one species to another Not single copy genes,repeated many times -Some copies are identical -Others are quite different -H4 has only had 2 variants ever reported 13-3
13-3 Properties of Histones ✓ Abundant proteins whose mass in nuclei nearly equals that of DNA ✓ Pronounced positive charge at neutral pH ✓ Most are well-conserved from one species to another ✓ Not single copy genes, repeated many times – Some copies are identical – Others are quite different – H4 has only had 2 variants ever reported

13.2 Nucleosomes(核小体) Chromosomes are long,thin molecules that will tangle(混乱状态)if not carefully folded Folding occurs in several ways First order of folding is the nucleosome -X-ray diffraction(衍射)has shown strong repeats of structure at 100A intervals -This spacing approximates the nucleosome spaced at 110A intervals 13-4
13-4 13.2 Nucleosomes (核小体) ✓ Chromosomes are long, thin molecules that will tangle (混乱状态) if not carefully folded ✓ Folding occurs in several ways ✓ First order of folding is the nucleosome – X-ray diffraction (衍射) has shown strong repeats of structure at 100Å intervals – This spacing approximates the nucleosome spaced at 110Å intervals
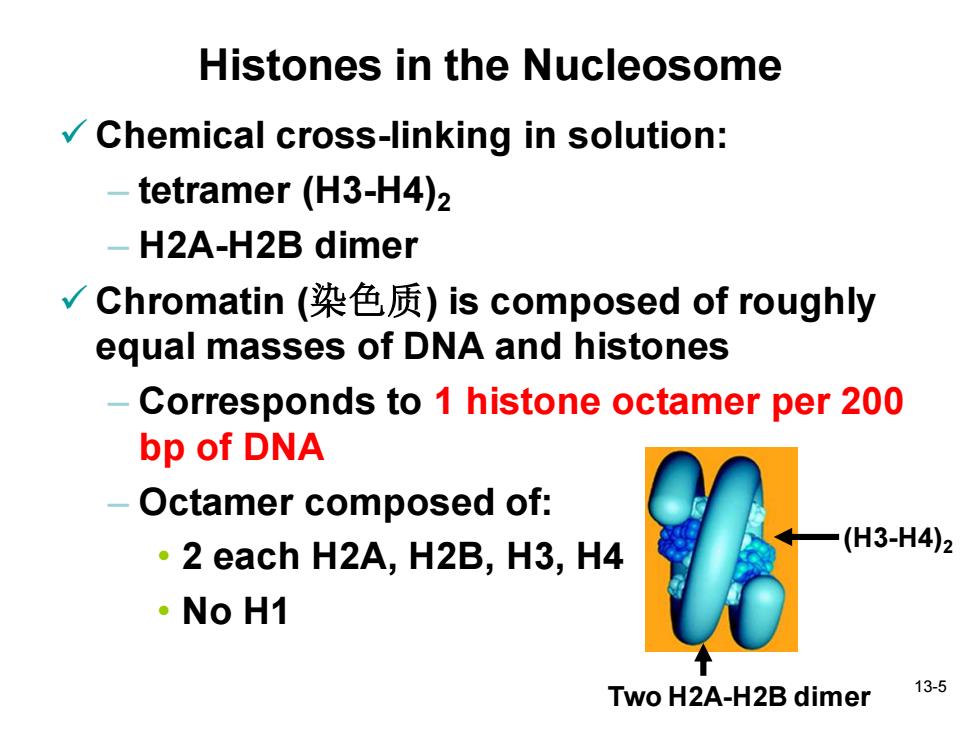
Histones in the Nucleosome Chemical cross-linking in solution: -tetramer(H3-H4)2 - H2A-H2B dimer √Chromatin(染色质)is composed of roughly equal masses of DNA and histones Corresponds to 1 histone octamer per 200 bp of DNA -Octamer composed of: ·2 each H2A,H2B,H3,H4 (H3-H4)2 ·NoH1 Two H2A-H2B dimer 13-5
13-5 Histones in the Nucleosome ✓ Chemical cross-linking in solution: – tetramer (H3-H4)2 – H2A-H2B dimer ✓ Chromatin (染色质) is composed of roughly equal masses of DNA and histones – Corresponds to 1 histone octamer per 200 bp of DNA – Octamer composed of: • 2 each H2A, H2B, H3, H4 • No H1 (H3-H4)2 Two H2A-H2B dimer
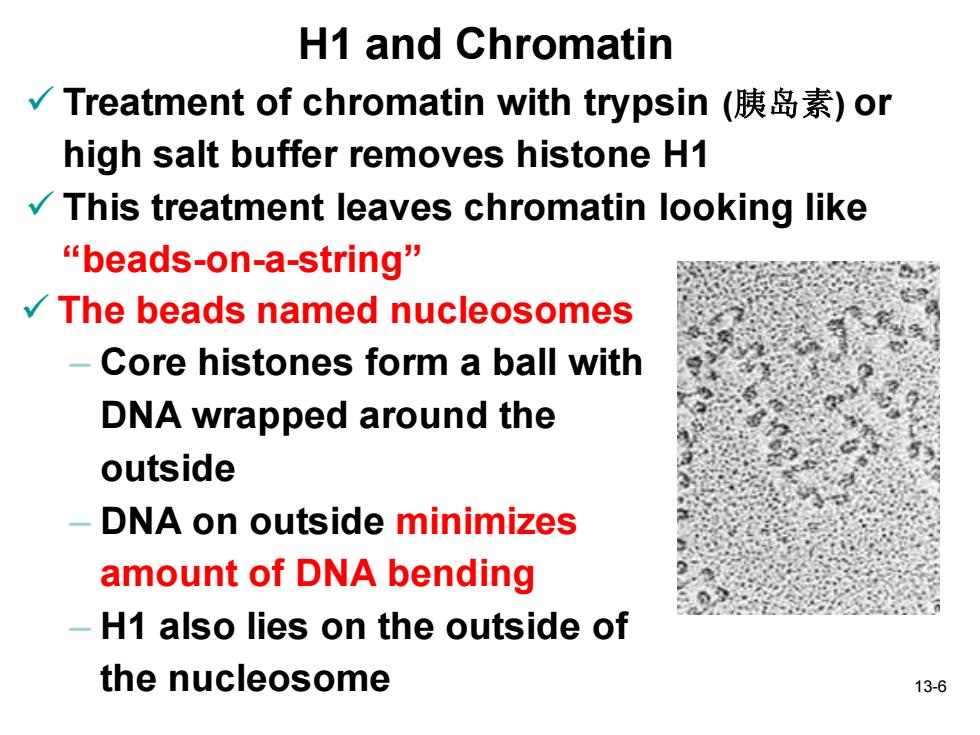
H1 and Chromatin √Treatment of chromatin with trypsin(胰岛素)or high salt buffer removes histone H1 This treatment leaves chromatin looking like beads-on-a-string” The beads named nucleosomes Core histones form a ball with DNA wrapped around the outside DNA on outside minimizes amount of DNA bending -H1 also lies on the outside of the nucleosome 13-6
13-6 H1 and Chromatin ✓ Treatment of chromatin with trypsin (胰岛素) or high salt buffer removes histone H1 ✓ This treatment leaves chromatin looking like “beads-on-a-string” ✓ The beads named nucleosomes – Core histones form a ball with DNA wrapped around the outside – DNA on outside minimizes amount of DNA bending – H1 also lies on the outside of the nucleosome
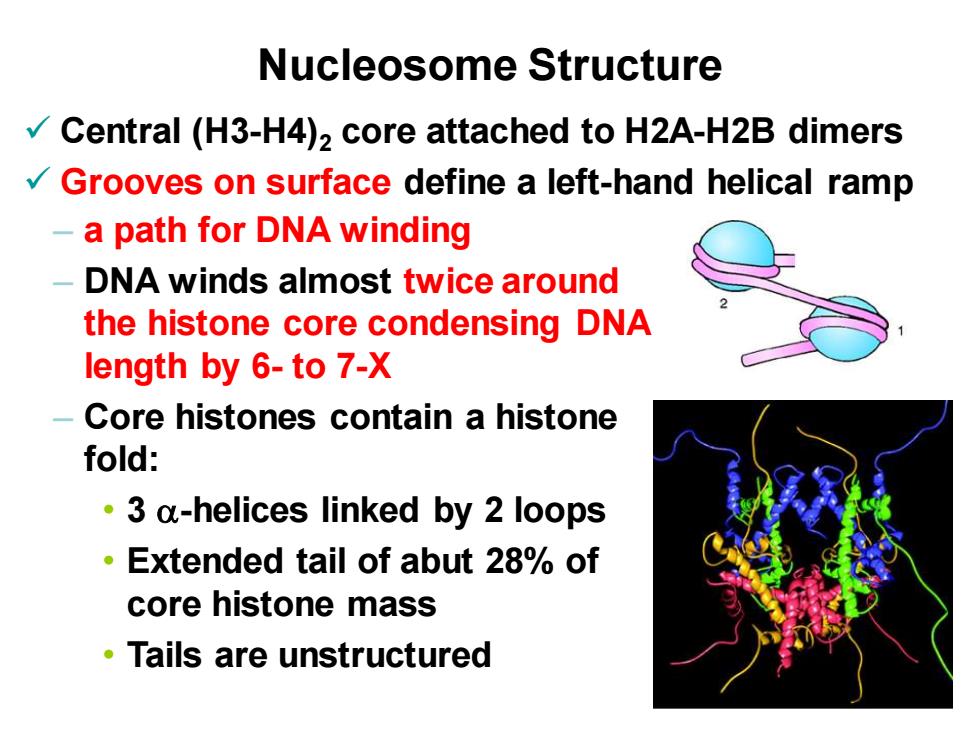
Nucleosome Structure Central(H3-H4)2 core attached to H2A-H2B dimers Grooves on surface define a left-hand helical ramp -a path for DNA winding DNA winds almost twice around the histone core condensing DNA length by 6-to 7-X Core histones contain a histone fold: 3 a-helices linked by 2 loops Extended tail of abut 28%of core histone mass Tails are unstructured
13-7 Nucleosome Structure ✓ Central (H3-H4)2 core attached to H2A-H2B dimers ✓ Grooves on surface define a left-hand helical ramp – a path for DNA winding – DNA winds almost twice around the histone core condensing DNA length by 6- to 7-X – Core histones contain a histone fold: • 3 a-helices linked by 2 loops • Extended tail of abut 28% of core histone mass • Tails are unstructured
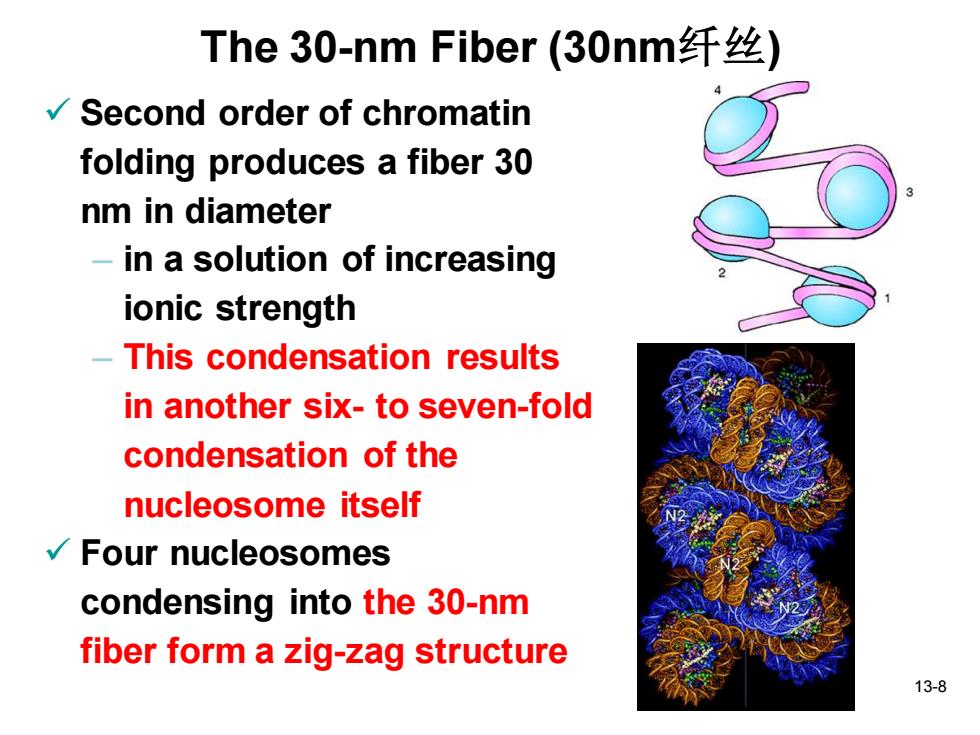
The30-nm Fiber(30nm纤丝) Second order of chromatin folding produces a fiber 30 nm in diameter -in a solution of increasing ionic strength This condensation results in another six-to seven-fold condensation of the nucleosome itself √Four nucleosomes condensing into the 30-nm fiber form a zig-zag structure 13-8
13-8 The 30-nm Fiber (30nm纤丝) ✓ Second order of chromatin folding produces a fiber 30 nm in diameter – in a solution of increasing ionic strength – This condensation results in another six- to seven-fold condensation of the nucleosome itself ✓ Four nucleosomes condensing into the 30-nm fiber form a zig-zag structure
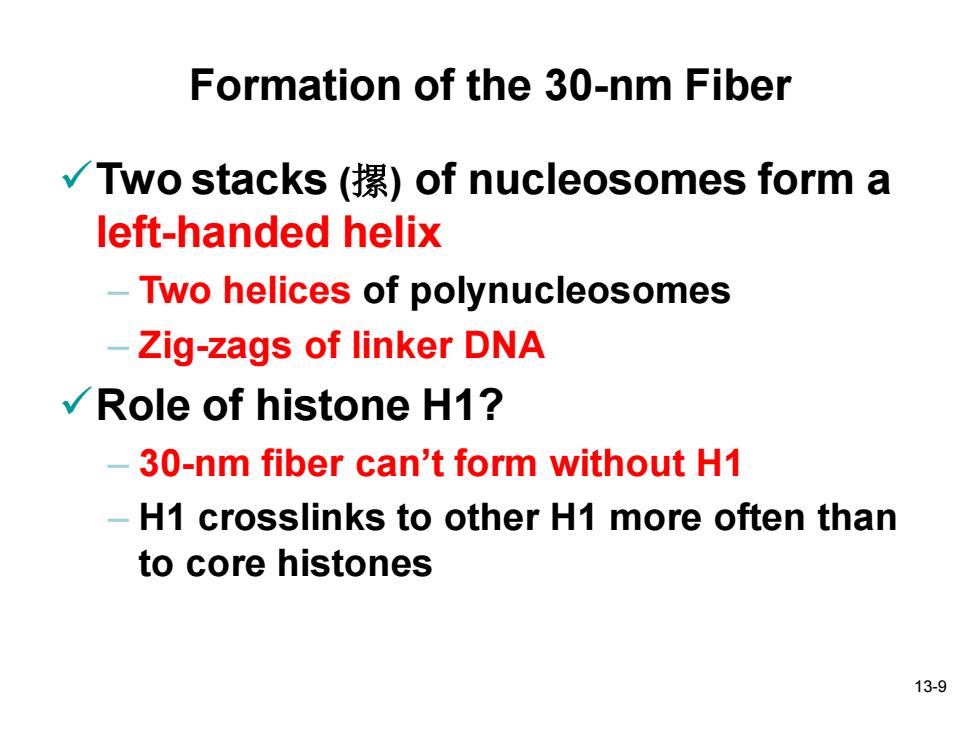
Formation of the 30-nm Fiber √Two stacks(摞)of nucleosomes form a left-handed helix -Two helices of polynucleosomes Zig-zags of linker DNA √/Role of histone H1? -30-nm fiber can't form without H1 -H1 crosslinks to other H1 more often than to core histones 13-9
13-9 Formation of the 30-nm Fiber ✓Two stacks (摞) of nucleosomes form a left-handed helix – Two helices of polynucleosomes – Zig-zags of linker DNA ✓Role of histone H1? – 30-nm fiber can’t form without H1 – H1 crosslinks to other H1 more often than to core histones
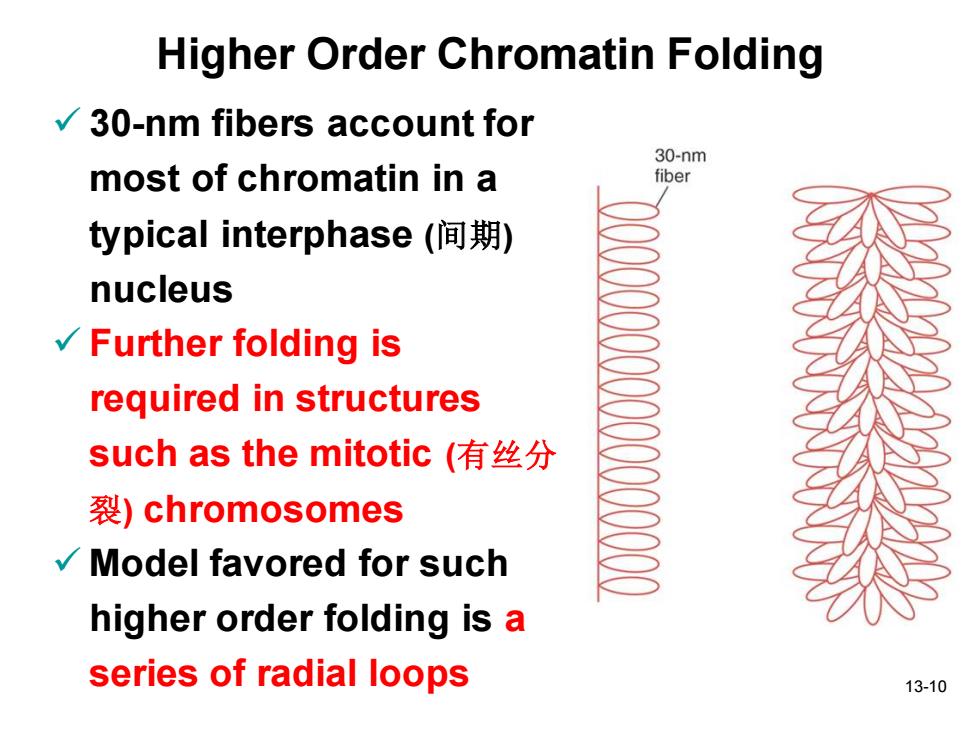
Higher Order Chromatin Folding 30-nm fibers account for 30-nm most of chromatin in a fiber typical interphase(间期) nucleus v√Further folding is required in structures such as the mitotic(有丝分 裂)chromosomes Model favored for such higher order folding is a series of radial loops 13-10
13-10 Higher Order Chromatin Folding ✓ 30-nm fibers account for most of chromatin in a typical interphase (间期) nucleus ✓ Further folding is required in structures such as the mitotic (有丝分 裂) chromosomes ✓ Model favored for such higher order folding is a series of radial loops