
Wireg Chen'4 Group of Bioinformatics @College of Life Science,Zhejiang University 0心转录组学000队 01/15,2017 ventson@zju.edu.cn
转录组学 01/15, 2017 ventson@zju.edu.cn Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University

Ting Chen'4 Group of Bioinformatics @College of Life Science,Zhejiang University 00八 前言 replication (DNA->DNA) DNA Polymerase 基因组学 QD0①d1 DNA transcription (DNA->RNA) RNA Polymerase RNA 转录组学 translation (RNA->Protein) Ribosome ○OOoO-Protein 蛋白质组学 from en.wikipedia 中心法则:遗传信息传递 2
前言 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University from en.wikipedia 基因组学 转录组学 蛋白质组学 中心法则:遗传信息传递 2
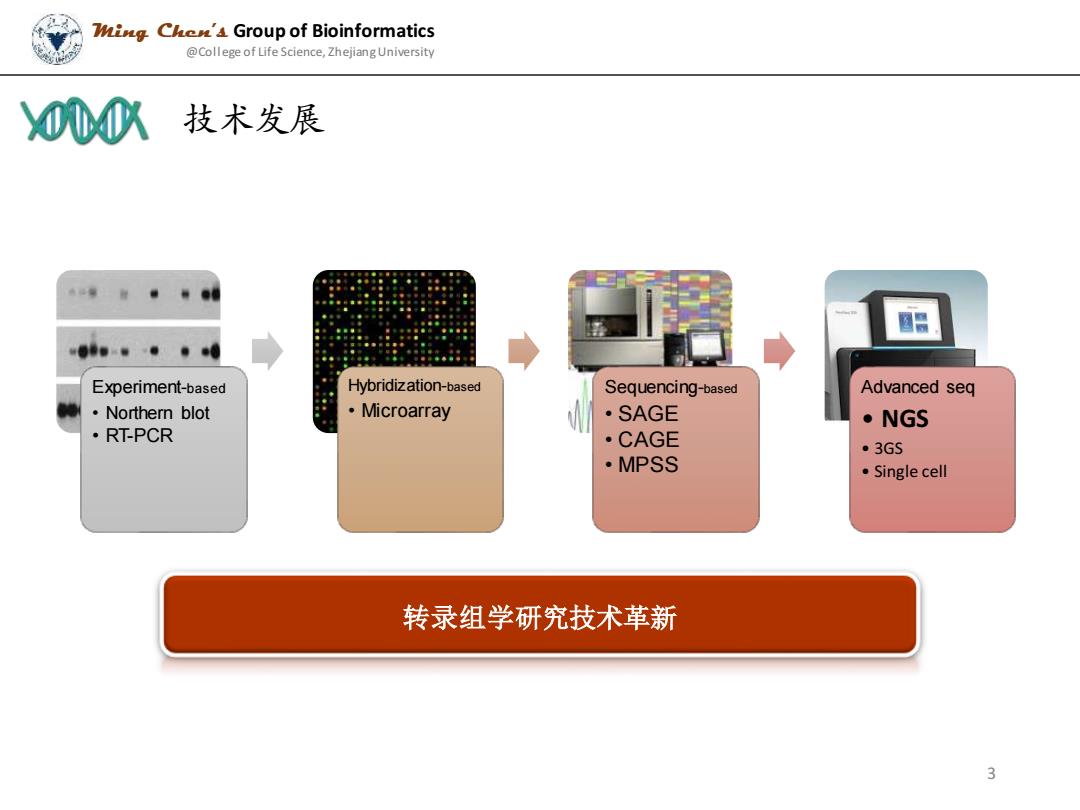
Zig Che'4 Group of Bioinformatics @College of Life Science,Zhejiang University 00心 技术发展 国日 Experiment-based Hybridization-based Sequencing-based Advanced seq ·Northern blot ·Microarray ·SAGE ·NGS ·RT-PCR ·CAGE ·3GS ·MPSS ·Single cell 转录组学研究技术革新 3
技术发展 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University Experiment-based • Northern blot • RT-PCR Hybridization-based • Microarray Sequencing-based • SAGE • CAGE • MPSS Advanced seq • NGS • 3GS • Single cell 转录组学研究技术革新 3
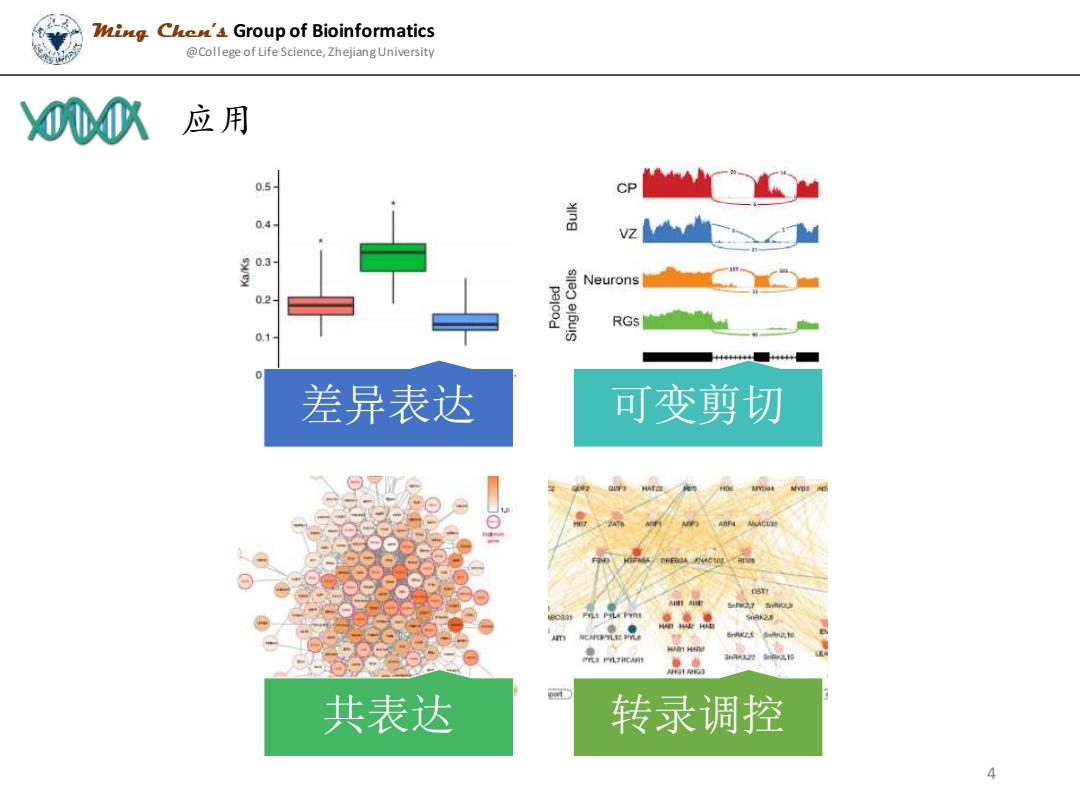
ing Chen'4 Group of Bioinformatics @College of Life Science,Zhejiang University 00八应用 0.5- CP 盖 0.3 Neurons 02 RGs 0.1 差异表达 可变剪切 共表达 转录调控 4
应用 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 差异表达 可变剪切 共表达 转录调控 4

WigChe'4 Group of Bioinformatics @College of Life Science,Zhejiang University O价RNA测序(RNA--sequencing) AARAAAA西 TTTI TGATAGATC Reverse transcriptase GGCGATCG TAGCTGTAC ARAAAAAA TTTT RNA CDNA Library Data isolation amplification Sequencing preparation analysis from GATC Biotech DNA 1.试验设计 2.测序流程 3.数据分析 4.验证实验 5
RNA测序(RNA-sequencing) Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 1.试验设计 2.测序流程 3.数据分析 4.验证实验 from GATC Biotech 5

WigChe'4 Group of Bioinformatics @College of Life Science,Zhejiang University 000八 试验设计 问题导向型 数据导向型 生物学重复(3-5个) 数据异质性(平台、个体差异) 样本提取(分类和保存) 确定分析流程 测序深度(简单基因表达分析需5M以 上reads,小RNA至少30M) 分析工具选用 文库构建(链特异性非特异性) 测序策略(单端和双末端) 测序平台(读长、通量和准确率等) 6
问题导向型 生物学重复(3-5个) 样本提取(分类和保存) 测序深度(简单基因表达分析需5M以 上reads,小RNA至少30M) 文库构建(链特异性非特异性) 测序策略(单端和双末端) 测序平台(读长、通量和准确率等) 数据导向型 数据异质性(平台、个体差异) 确定分析流程 分析工具选用 试验设计 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 6

Ting Chen'4 Group of Bioinformatics @College of Life Science,Zhejiang University 00八 测序流程 目标样本 RNA分离纯化 打断,构建cDNA文库, MMMMM 长度筛选,添加接头 AuAuAwAAuAAAAuuA 肿瘤组织 正常组织 M mRNA:Poly A富集Poly(A)尾 ncRNA:rRNA移除 上机测序 比对到参考基因组或转录组 内含子 RNA前体 外显子 未测序RNA RNA读段 M 转录本 短读段 可变剪切区域 短插入片段 Griffith,M.(2015)PLoS computational biology 7
Griffith, M. (2015) PLoS computational biology mRNA:Poly A富集 ncRNA:rRNA移除 测序流程 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 7
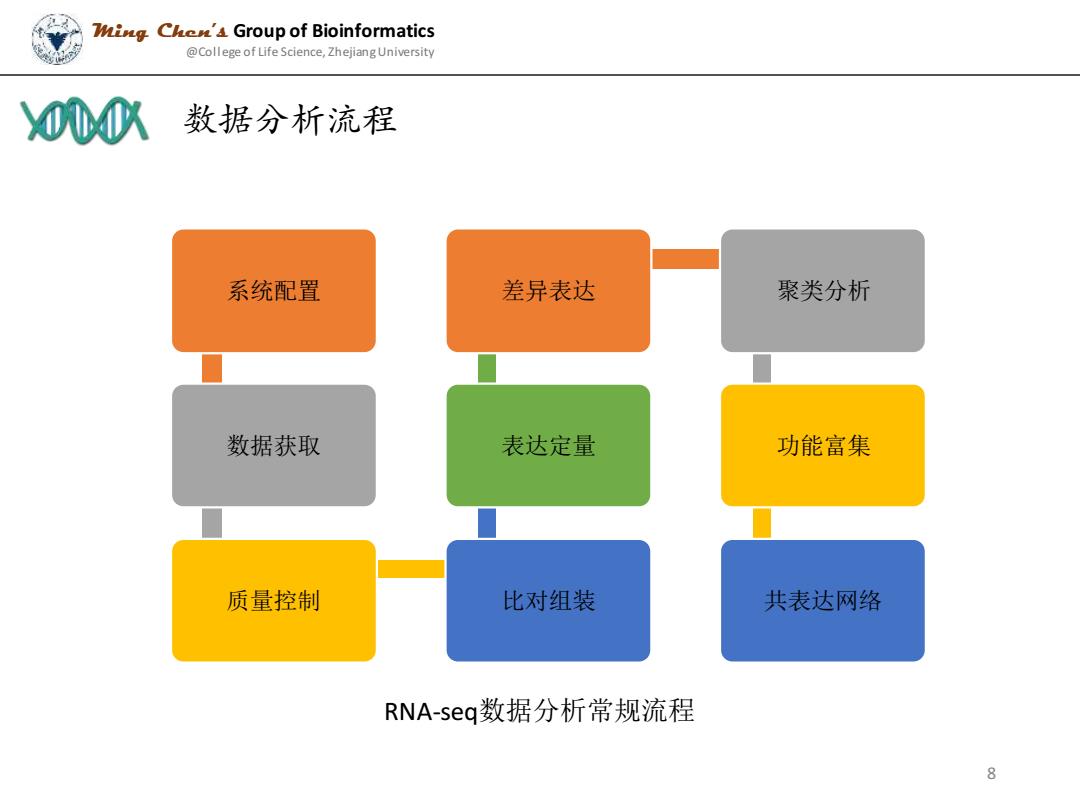
Wing Che'4 Group of Bioinformatics @College of Life Science,Zhejiang University 000队 数据分析流程 系统配置 差异表达 聚类分析 数据获取 表达定量 功能富集 质量控制 比对组装 共表达网络 RNA-seq数据分析常规流程 8
系统配置 数据获取 质量控制 比对组装 表达定量 差异表达 聚类分析 功能富集 共表达网络 RNA-seq数据分析常规流程 数据分析流程 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 8

Zing Chen'4 Group of Bioinformatics @College of Life Science,Zhejiang University 00 、系统配置 8L△nux Microsoft Java Sun Microsystems R 语言基础 080+ Public Galaxy Servers and stiff counting 9
系统配置 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 9

WigChe'4 Group of Bioinformatics @College of Life Science,Zhejiang University 000队 数据获取 GSA THE CANCER GENOME ATLAS NIH National Cancer Institute National Human Genome Research Institute Genome Sequence Archive NCBI SRA TCGA/GDC(cancer) ArrayExpress NIH NATIONAL CANCER INSTITUTE Genomic Data Commons fastq-dump EBI ArrayExpress (SRAToolkit) 公共数据库 测序公司 Fastq文件格式: esRR3418005.1HAL:1282:D2 EWTACXX:8:1101:1602:21361 ength=100 GGCAAGATCTGATCTCTCAGCAACTCAATTACAACCATAACCGCGTGTGACTTCTAAGCC +sRR3418005.1HAL:1282:D2 EWTACXX:8:1101:1602:21361 ength=100 :1DDAD?DDFHFGHIIGEIIIGHHHDHGIGDFGHGGEGIIHGBEDHEEGGFIDEEAAEH @3RR3418005,2HAL:1282:D2 EWTACXX:8:1101:1550:21931 ength-100 ATCTGATTCAATCATAAATTTTACACAATCAATTTGTCGGTACTCTCCTTTTGGTCATAT +SRR3418005.2HAL:1282:D2 EWTACXX:8:1101:1550:21931 ength-100 <?0BDD:DCDDHHGIBGA<FFF<:FDGGCE9CADHTEG:?DF):BBB9??BGG60?B<B- esRR3418005.3HAL:1282:D2 EWTACXX:8:1101:1632:22051 ength=100 AGACGCTCGTACCAAATCCGTTACCGTCTCCGTCGTTACCTCCTCCTTCGCGACGGGAAC +sRR3418005.3HAL:1282:D2 EWTACXX:8:1101:1632:22051 ength-100 --+:A0DDF0F8C<A+CBEFIIFIFODFFFFFDF(0F--.-70FEF).-0A# esRR3418005.4HAL:1282:D2 EWTACXX:8:1101:1588:22271 ength-100 CTCATTTTTATTACCGCATATATGACATATGATCAATTACATAAAGAAGCAAATCTTAGO +SRR3418005.4HAL:1282:D2 EWTACXX:8:1101:1588:22271 ength-100 G-DDDAFHFAHEH?FF<EBF9EHDHHGOACCGICHHCHGIEHG<DFB9DDFHEGFHDG 8sRR3418005,5HAL:1282:D2 EWTACXX:8:1101:1991:21131 ength=100 CGGAAGCAGCTGAGAAGCCTCATGGTTACCAACAAGAGCATCCTCATCAGTTNCACCATA +sRR3418005,5HAL:1282:D2 EWTACXX:8:1101:1991:21131 ength-100 BCOFFFDFHHHFGIIGIGIJJJJIJIIIIIIJJJJCEIGIIJHIJEIIJJHI-<FFHIJ 10
Fastq文件格式: NCBI SRA EBI ArrayExpress TCGA/GDC(cancer) fastq-dump (SRAToolkit) 公共数据库 测序公司 数据获取 Ming Chen’s Group of Bioinformatics @College of Life Science, Zhejiang University 10